Abstract
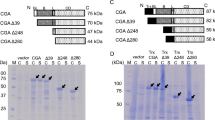
Thermoanaerobacter tengcongensis MB4 glucoamylase (TteGA) contains a catalytic domain (CD), which is structurally similar to eukaryotic GA, and a β domain (BD) with ambiguous function. Firstly, BD is found to be essential to TteGA activity because CD alone could not hydrolyze soluble starch. However, starch hydrolysis activity, similar to that of intact TteGA, was restored to CD in the presence of BD. Secondly, BD is found to be an important helper in the correct folding of CD because CD was mainly expressed in the inclusion bodies on its own in Escherichia coli. By contrast, intact TteGA, BD, and CD combined with BD could be expressed as soluble proteins. Additionally, BD is essential to the thermostability of TteGA because CD displayed lower thermostability compared with the intact TteGA and exhibited enhanced thermostability in the presence of BD in vitro. Truncation of TteGA or mutagenesis of the residues that participate in the interdomain interaction at its BD also led to the reduced thermostability of TteGA.





Similar content being viewed by others
References
Aleshin AE, Feng PH, Honzatko RB, Reilly PJ (2003) Crystal structure and evolution of a prokaryotic glucoamylase. J Mol Biol 327:61–73
Allen MJ, Coutinho PM, Ford CF (1998) Stabilization of Aspergillus awamori glucoamylase by proline substitution and combining stabilizing mutations. Protein Eng 11:783–788
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Bhaskara RM, Srinivasan N (2011) Stability of domain structures in multi-domain proteins. Sci Rep 1:1–9
Bott R, Saldajeno M, Cuevas W, Ward D, Scheffers M, Aehle W, Karkehabadi S, Sandgren M, Hansson H (2008) Three-dimensional structure of an intact glycoside hydrolase family 15 glucoamylase from Hypocrea jecorina. Biochemistry 47:5746–5754
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Chen HM, Bakir U, Reilly PJ, Ford C (1994) Increased thermostability of Asn182 –> Ala mutant Aspergillus awamori glucoamylase. Biotechnol Bioengg 43:101–105
Cheng HR, Jiang N (2006) Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett 28:55–59
Coutinho PM, Reilly PJ (1997) Glucoamylase structural, functional, and evolutionary relationships. Proteins 29:334–347
Dock C, Hess M, Antranikian G (2008) A thermoactive glucoamylase with biotechnological relevance from the thermoacidophilic Euryarchaeon Thermoplasma acidophilum. Appl Microbiol Biotechnol 78:105–114
Ducki A, Grundmann O, Konermann L, Mayer F, Hoppert M (1998) Glucoamylase from Thermoanaerobacterium thermosaccharolyticum: sequence studies and analysis of the macromolecular architecture of the enzyme. J Gen Appl Microbiol 44:327–335
Feng DR (1999) Biosensors and their application in the People’s Republic of China. Advances in Biosensors 4:289–313
Ford C (1999) Improving operating performance of glucoamylase by mutagenesis. Curr Opin Biotechnol 10:353–357
Gill RK, Kaur J (2004) A thermostable glucoamylase from a thermophilic Bacillus sp.: characterization and thermostability. J Ind Microbiol Biotechnol 31:540–543
Golubev AM, Neustroev KN, Aleshin AE, Firsov LM (1992) Crystallization and preliminary X-ray study of minor glucoamylase from Aspergillus awamori variant X-100/D27. J Mol Biol 226:271–272
Henrissat B, Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 293:781–788
Horvathova V, Slajsova K, Sturdik E (2004) Evaluation of the glucoamylase Glm from Saccharomycopsis fibuligera IFO 0111 in hydrolysing the corn starch. Biologia 59:361–365
Hostinová E, Gašperík J (2010) Yeast glucoamylases: molecular-genetic and structural characterization. Biologia 65:559–568
Hostinova E (2002) Amylolytic enzymes produced by the yeast Saccharomycopsis fibuligera. Biologia 57:247–251
Hostinova E, Solovicova A, Dvorsky R, Gasperik J (2003) Molecular cloning and 3D structure prediction of the first raw-starch-degrading glucoamylase without a separate starch-binding domain. Arch Biochem Biophys 411:189–195
Jorgensen AD, Nohr J, Kastrup JS, Gajhede M, Sigurskjold BW, Sauer J, Svergun DI, Svensson B, Vestergaard B (2008) Small angle X-ray studies reveal that Aspergillus niger glucoamylase has a defined extended conformation and can form dimers in solution. J Biol Chem 283:14772–14780
Kelley LA, Sternberg MJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371
Kim MS, Park JT, Kim YW, Lee HS, Nyawira R, Shin HS, Park CS, Yoo SH, Kim YR, Moon TW, Park KH (2004) Properties of a novel thermostable glucoamylase from the hyperthermophilic archaeon Sulfolobus solfataricus in relation to starch processing. Appl Environ Microbiol 70:3933–3940
Kumar P, Satyanarayana T (2009) Microbial glucoamylases: characteristics and applications. Crit Rev Biotechnol 29:225–255
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Li Y, Coutinho PM, Ford C (1998) Effect on thermostability and catalytic activity of introducing disulfide bonds into Aspergillus awamori glucoamylase. Protein Eng 11:661–667
Libby CB, Cornett CA, Reilly PJ, Ford C (1994) Effect of amino acid deletions in the O-glycosylated region of Aspergillus awamori glucoamylase. Protein Eng 7:1109–1114
Liu HL, Wang WC (2003) Protein engineering to improve the thermostability of glucoamylase from Aspergillus awamori based on molecular dynamics simulations. Protein Eng 16:19–25
McDaniel A, Fuchs E, Liu Y, Ford C (2008) Directed evolution of Aspergillus niger glucoamylase to increase thermostability. Microb Biotechnol 1:523–531
Mertens JA, Skory CD (2007) Isolation and characterization of two genes that encode active glucoamylase without a starch binding domain from Rhizopus oryzae. Curr Microbiol 54:462–466
Norouzian D, Akbarzadeh A, Scharer JM, Young MM (2006) Fungal glucoamylases. Biotechnol Adv 24:80–85
Ohnishi H, Kitamura H, Minowa T, Sakai H, Ohta T (1992) Molecular cloning of a glucoamylase gene from a thermophilic Clostridium and kinetics of the cloned enzyme. Eur J Biochem 207:413–418
Paldi T, Levy I, Shoseyov O (2003) Glucoamylase starch-binding domain of Aspergillus niger B1: molecular cloning and functional characterization. Biochem J 372:905–910
Rodriguez-Sanoja R, Oviedo N, Sanchez S (2005) Microbial starch-binding domain. Curr Opin Microbiol 8:260–267
Sakai Y, Akiyama M, Kondoh H, Shibano Y, Kato N (1996) High-level secretion of fungal glucoamylase using the Candida boidinii gene expression system. Biochim Biophys Acta 1308:81–87
Sevcik J, Hostinova E, Solovicova A, Gasperik J, Dauter Z, Wilson KS (2006) Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain. FEBS J 273:2161–2171
Sevcik J, Solovicova A, Hostinova E, Gasperik J, Wilson KS, Dauter Z (1998) Structure of glucoamylase from Saccharomycopsis fibuligera at 1.7 A resolution. Acta Crystallogr D: Biol Crystallogr 54:854–866
Sorimachi K, Jacks AJ, Le Gal-Coeffet MF, Williamson G, Archer DB, Williamson MP (1996) Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy. J Mol Biol 259:970–987
Specka U, Mayer F, Antranikian G (1991) Purification and properties of a thermoactive glucoamylase from Clostridium thermosaccharolyticum. Appl Environ Microbiol 57:2317–2323
Stamford TL, Stamford NP, Coelho LC, Araujo JM (2002) Production and characterization of a thermostable glucoamylase from Streptosporangium sp. endophyte of maize leaves. Biores Technol 83:105–109
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tung JY, Chang MD, Chou WI, Liu YY, Yeh YH, Chang FY, Lin SC, Qiu ZL, Sun YJ (2008) Crystal structures of the starch-binding domain from Rhizopus oryzae glucoamylase reveal a polysaccharide-binding path. Biochem J 416:27–36
Uotsu-Tomita R, Tonozuka T, Sakai H, Sakano Y (2001) Novel glucoamylase-type enzymes from Thermoactinomyces vulgaris and Methanococcus jannaschii whose genes are found in the flanking region of the alpha-amylase genes. Appl Microbiol Biotechnol 56:465–473
Xue YF, Xu Y, Liu Y, Ma YH, Zhou PJ (2001) Thermoanaerobacter tengcongensis sp nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int J Syst Evol Microbiol 51:1335–1341
Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE (2012) UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol 179:269–278
Zheng Y, Xue Y, Zhang Y, Zhou C, Schwaneberg U, Ma Y (2010) Cloning, expression, and characterization of a thermostable glucoamylase from Thermoanaerobacter tengcongensis MB4. Appl Microbiol Biotechnol 87:225–233
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (863 Program, 2012AA022203), and the National Basic Research Program (973 Program, 2011CBA00800). Q.W. is supported by the Bairenjihhua Program of the Chinese Academy of Sciences. The authors are grateful to Prof. Keqian Yang for comments and discussions of this paper, and Dr. Ence Yang for critically reading this paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1836 kb)
Rights and permissions
About this article
Cite this article
Li, Z., Wei, P., Cheng, H. et al. Functional role of β domain in the Thermoanaerobacter tengcongensis glucoamylase. Appl Microbiol Biotechnol 98, 2091–2099 (2014). https://doi.org/10.1007/s00253-013-5051-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5051-2