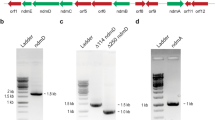
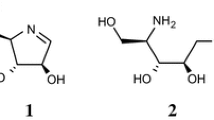
Abstract
Cupriavidus necator JMP134 utilizes meta-nitrophenol (MNP) as a sole source of carbon, nitrogen, and energy. The metabolic reconstruction of MNP degradation performed in silico suggested that the mnp cluster might have played important roles in MNP degradation. In order to experimentally confirm the prediction, we have now characterized mnpA-encoded meta-nitrophenol nitroreductase involved in the initial reaction of MNP degradation. Real-time PCR analysis indicated that mnpA played an essential role in MNP degradation. MnpA was purified to homogeneity as His-tagged proteins and was considered to be a dimer as determined by gel filtration. MnpA was an MNP nitroreductase with a tightly bound flavin mononucleotide (FMN), catalyzing the partial reduction of MNP to meta-hydroxylaminophenol via meta-nitrosophenol in the presence of NADPH and oxygen. The accumulation of meta-nitrosophenol was confirmed with the results of liquid chromatography–diode array detection and time-of-flight mass spectrometry for the first time. The low Km and high kcat of MnpA as well as MNP-inducible transcription of mnpA suggested that MNP was the physiological substrate for this nitroreductase. In addition, the phylogenetic analysis revealed that nitroreductases of known physiological function including MnpA constituted a new clade in the nitro-FMN-reductase superfamily.




Similar content being viewed by others
References
Becker AR, Sternson LA (1980) Non-enzymatic reduction of nitrosobenzene to phenylhydroxylamine by NAD(P)H. Bioorg Chem 9:305–312
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bruhn C, Lenke H, Knackmuss HJ (1987) Nitrosubstituted aromatic-compounds as nitrogen-source for bacteria. Appl Environ Microbiol 53:208–210
Do CB, Mahabhashyam MS, Brudno M, Batzoglou S (2005) ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res 15:330–340
Groenewegen PE, Breeuwer P, van Helvoort JM, Langenhoff AA, de Vries FP, de Bont JA (1992) Novel degradative pathway of 4-nitrobenzoate in Comamonas acidovorans NBA-10. J Gen Microbiol 138:1599–1605
Hughes MA, Williams PA (2001) Cloning and characterization of the pnb genes, encoding enzymes for 4-nitrobenzoate catabolism in Pseudomonas putida TW3. J Bacteriol 183:1225–1232
Iwaki H, Muraki T, Ishihara S, Hasegawa Y, Rankin KN, Sulea T, Boyd J, Lau PCK (2007) Characterization of a pseudomonad 2-nitrobenzoate nitroreductase and its catabolic pathway-associated 2-hydroxylaminobenzoate mutase and a chemoreceptor involved in 2-nitrobenzoate chemotaxis. J Bacteriol 189:3502–3514
Jain RK, Dreisbach JH, Spain JC (1994) Biodegradation of p-nitrophenol via 1, 2, 4-benzenetriol by an Arthrobacter sp. Appl Environ Microbiol 60:3030–3032
Kitagawa W, Kimura N, Kamagata Y (2004) A novel p-nitrophenol degradation gene cluster from a Gram-positive bacterium, Rhodococcus opacus SAO101. J Bacteriol 186:4894–4902
Kuhm AE, Schlomann M, Knackmuss HJ, Pieper DH (1990) Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP134. Biochem J 266:877–883
Leskovac V, Svircevic J, Trivic S, Popovic M, Radulovic M (1989) Reduction of aryl nitroso-compounds by pyridine and flavin coenzymes. Int J Biochem 21:825–834
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Louie TM, Webster CM, Xun LY (2002) Genetic and biochemical characterization of a 2, 4, 6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134. J Bacteriol 184:3492–3500
Meulenberg R, Pepi M, de Bont JA (1996) Degradation of 3-nitrophenol by Pseudomonas putida B2 occurs via 1, 2, 4-benzenetriol. Biodegradation 7:303–311
Ooi T, Shibata T, Sato R, Ohno H, Kinoshita S, Thuoc TL, Taguchi S (2007) An azoreductase, aerobic NADH-dependent flavoprotein discovered from Bacillus sp.: functional expression and enzymatic characterization. Appl Environ Microbiol 75:377–386
Park HS, Kim HS (2000) Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J Bacteriol 182:573–580
Perez-Pantoja D, De la Iglesia R, Pieper DH, Gonzalez B (2008) Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol Rev 32:736–794
Perez-Reinado E, Blasco R, Castillo F, Moreno-Vivian C, Roldan MD (2005) Regulation and characterization of two nitroreductase genes, nprA and nprB, of Rhodobacter capsulatus. Appl Environ Microbiol 71:7643–7649
Race PR, Lovering AL, Green RM, Ossor A, White SA, Searle PF, Wrighton CJ, Hyde EI (2005) Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme. J Biol Chem 280:13256–13264
Rau J, Stolz A (2003) Oxygen-insensitive nitroreductases NfsA and NfsB of Escherichia coli function under anaerobic conditions as lawsone-dependent azo reductases. Appl Environ Microbiol 69:3448–3455
Rieger PG, Meier HM, Gerle M, Vogt U, Groth T, Knackmuss HJ (2002) Xenobiotics in the environment: present and future strategies to obviate the problem of biological persistence. J Biotechnol 94:101–123
Roldan MD, Perez-Reinado E, Castillo F, Moreno-Vivian C (2008) Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol Rev 32:474–500
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schenzle A, Lenke H, Fischer P, Williams PA, Knackmuss H (1997) Catabolism of 3-nitrophenol by Ralstonia eutropha JMP134. Appl Environ Microbiol 63:1421–1427
Schenzle A, Lenke H, Spain JC, Knackmuss HJ (1999a) 3-Hydroxylaminophenol mutase from Ralstonia eutropha JMP134 catalyzes a Bamberger rearrangement. J Bacteriol 181:1444–1450
Schenzle A, Lenke H, Spain JC, Knackmuss HJ (1999b) Chemoselective nitro group reduction and reductive dechlorination initiate degradation of 2-chloro-5-nitrophenol by Ralstonia eutropha JMP134. Appl Environ Microbiol 65:2317–2323
Somerville CC, Nishino SF, Spain JC (1995) Purification and characterization of nitrobenzene nitroreductase from Pseudomonas pseudoalcaligenes JS45. J Bacteriol 177:3837–3842
Spain JC, Wyss O, Gibson DT (1979) Enzymatic oxidation of p-nitrophenol. Biochem Biophys Res Commun 88:634–641
Streker K, Freiberg C, Labischinski H, Hacker J, Ohlsen K (2005) Staphylococcus aureus NfrA (SA0367) is a flavin mononucleotide-dependent NADPH oxidase involved in oxidative stress response. J Bacteriol 187:2249–2256
Timmis KN, Pieper DH (1999) Bacteria designed for bioremediation. Trends Biotechnol 17:201–204
Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert IB (1998) Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol 180:5529–5539
Wu JF, Jiang CY, Wang BJ, Ma YF, Liu ZP, Liu SJ (2006) Novel partial reductive pathway for 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB1. Appl Environ Microbiol 72:1759–1765
Xiao Y, Wu JF, Liu H, Wang SJ, Liu SJ, Zhou NY (2006) Characterization of genes involved in the initial reactions of 4-chloronitrobenzene degradation in Pseudomonas putida ZWL73. Appl Microbiol Biotechnol 73:166–171
Xiao Y, Zhang JJ, Liu H, Zhou NY (2007) Molecular characterization of a novel ortho-nitrophenol catabolic gene cluster in Alcaligenes sp. strain NyZ215. J Bacteriol 189:6587–6593
Zeyer J, Kearney PC (1984) Degradation of o-nitrophenol and m-nitrophenol by a Pseudomonas putida. J Agric Food Chem 32:238–242
Zeyer J, Kocher HP (1988) Purification and characterization of a bacterial nitrophenol oxygenase which converts ortho-nitrophenol to catechol and nitrite. J Bacteriol 170:1789–1794
Zhang JJ, Liu H, Xiao Y, Zhang XE, Zhou NY (2009) Identification and characterization of catabolic para-nitrophenol 4-monooxygenase and para-benzoquinone reductase from Pseudomonas sp. strain WBC-3. J Bacteriol 191:2703–2710
Zhen D, Liu H, Wang SJ, Zhang JJ, Zhao F, Zhou NY (2006) Plasmid-mediated degradation of 4-chloronitrobenzene by newly isolated Pseudomonas putida ZWL73. Appl Microbiol Biotechnol 72:797–803
Zhou NY, Fuenmayor SL, Williams PA (2001) nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J Bacteriol 183:700–708
Acknowledgments
We acknowledge the financial supports from the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-YW-G-072-3) and National Natural Science Foundation of China (30730002 and 20825520).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yin, Y., Xiao, Y., Liu, HZ. et al. Characterization of catabolic meta-nitrophenol nitroreductase from Cupriavidus necator JMP134. Appl Microbiol Biotechnol 87, 2077–2085 (2010). https://doi.org/10.1007/s00253-010-2666-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2666-4