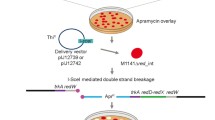
Abstract
Actinomycetes are Gram-positive bacteria with a complex life cycle. They produce many pharmaceutically relevant secondary metabolites, including antibiotics and anticancer drugs. However, there is a limited number of biotechnological applications available as opposed to genetic model organisms like Bacillus subtilis or Escherichia coli. We report here a system for the functional expression of a synthetic gene encoding the I-SceI homing endonuclease in several streptomycetes. Using the synthetic sce(a) gene, we were able to create controlled genomic DNA double-strand breaks. A mutagenesis system, based on the homing endonuclease I-SceI, has been developed to construct targeted, non-polar, unmarked gene mutations in Streptomyces sp. Tü6071. In addition, we have shown that homologous recombination is a major pathway in streptomycetes to repair an I-SceI-generated DNA double-strand break. This novel I-SceI-based tool will be useful in fundamental studies on the repair mechanism of DNA double-strand breaks and for a variety of biotechnological applications.



Similar content being viewed by others
References
Belfort M, Roberts RJ (1997) Homing endonucleases: keeping the house in order. Nucleic Acids Res 25:3379–3388
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood D (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bierman M, Logan R, O’Brian K, Seno ET, Rao RN, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116(1):43–49
Bihlmaier C (2005) Molekularbiologische Untersuchungen zur Biosynthese von Simoncyclinon und α-Lipomycin. Dissertation Albert-Ludwigs Universität Freiburg
Chevalier BS, Stoddard BL (2001) Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res 29:3757–3774
Choulika A, Perrin A, Dujon B, Nicolas JF (1995) Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol 15:1968–1973
Dujon B, Colleaux L, Jacquier A, Michel F, Monteilhet C (1986) Mitochondrial introns as mobile genetic elements: the role of intron-encoded proteins. Basic Life Sci 40:5–27
Dürr C, Schnell HJ, Luzhetskyy A, Murillo R, Weber M, Welzel K, Vente A, Bechthold A (2006) Biosynthesis of the terpene phenalinolactone in Streptomyces sp. Tü6071: analysis of the gene cluster and generation of derivatives. Chem Biol 13(4):365–377
Fedoryshyn M, Petzke L, Welle E, Bechthold A, Luzhetskyy A (2008) Marker removal from actinomycetes genome using FLP recombinase. Gene 419:43–47
Grabher C, Wittbrodt J (2008) Recent advances in meganuclease-and transposon-mediated transgenesis of medaka and zebrafish. Methods Mol Biol 461:521–539
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100:1541–1546
Haber JE (1995) In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays 17:609–620
Hesketh A, Bucca G, Laing E, Flett F, Hotchkiss G, Smith CP, Chater KF (2007) New pleiotropic effects of eliminating a rare tRNA from Streptomyces coelicolor, revealed by combined proteomic and transcriptomic analysis of liquid cultures. BMC Genomics 8:261–283
Huang TW, Chen CW (2006) A recA null mutant may be generated in Streptomyces coelicolor. J Bacteriol 188(19):6771–6779
Hütter R (1967) Systematik der Streptomyceten. Bibl Microbiol, 6, S. Karger, Basel, New York
Jacquier A, Dujon B (1985) An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 41:383–394
Janes BK, Stibitz S (2006) Routine markerless gene replacement in Bacillus anthracis. Infect Immun 74:1949–1953
Jasin M (1996) Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet 12:224–228
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Strepromyces genetics. John Innes Foundation, Norwich, United Kingdom
Lopez C, Rholl D, Trunk L, Schweizer P (2009) Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl Environ Microbiol 20:6496–6503
Luzhetskyy A, Fedoryshyn M, Gromyko O, Ostash B, Rebets Y, Bechthold A, Fedorenko V (2006) IncP plasmids are most effective in mediating conjugation between Escherichia coli and streptomycetes. Genetika 42:595–601
McEwan AR, Rowley PA, Smith MC (2009) DNA binding and synapsis by the large C-terminal domain of phiC31 integrase. Nucleic Acids Res 37:4764–4773
Moeller R, Stackebrandt E, Günther R, Berger T, Rettberg P, Doherty A, Horneck G, Nicholson W (2007) Role of DNA repair by nonhomologous-end joining in Bacillus subtilis spore resistance to extreme dryness, mono- and polychromatic UV, and ionizing radiation. J Bacteriol 189:3306–3311
Monteilhet C, Perrin A, Thierry A, Colleaux L, Dujon B (1990) Purification and characterization of the in vitro activity of I-Sce I, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res 18:1407–1413
Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ (1999) Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J Bacteriol 181:204–211
Perrin A, Buckle M, Dujon B (1993) Asymmetrical recognition and activity of the I-SceI endonuclease on its site and on intron–exon junctions. EMBO J 12:2939–2947
Petzke L, Luzhetskyy A (2008) In vivo Tn5 based transposon mutagenesis of Streptomycetes. Appl Microbiol Biotechnol 5:979–986
Posfai G, Kolisnychenko V, Bereczki Z, Blattner FR (1999) Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res 27:4409–4415
Rouet P, Smih F, Jasin M (1994) Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14:8096–8106
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M (1992) Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA 89(18):8794–8797
Smithies O (2001) Forty years with homologous recombination. Nat Med 7:1083–1086
Stephanou N, Gao F, Bongiorno P, Ehrt S, Schnappinger D, Shuman S, Glickman M (2007) Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J Bacteriol 189:5237–5246
Suzuki N, Nonaka H, Tsuge Y, Okayama S, Inui M, Yukawa H (2005) Multiple large segment deletion method for Corynebacterium glutamicum. Appl Microbiol Biotechnol 69:151–161
Wong SM, Mekalanos JJ (2000) Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 97:10191–10196
Acknowledgment
We are very grateful to Prof. A. Bechthold for critical discussions during the work as well as for the financial support. The authors also wish to thank Dr. B. Gust for providing plasmids pIJ778 and pIJ773, as well as Dr. M. Daum and K. Dietmann for the help in plasmid construction.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
(PDF 362 kb)
Rights and permissions
About this article
Cite this article
Siegl, T., Petzke, L., Welle, E. et al. I-SceI endonuclease: a new tool for DNA repair studies and genetic manipulations in streptomycetes. Appl Microbiol Biotechnol 87, 1525–1532 (2010). https://doi.org/10.1007/s00253-010-2643-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2643-y