Abstract
Recent progress has been made in the production of higher alcohols by harnessing the power of natural amino acid biosynthetic pathways. Here, we describe the first strain of Escherichia coli developed to produce the higher alcohol and potential new biofuel 2-methyl-1-butanol (2MB). To accomplish this, we explored the biodiversity of enzymes catalyzing key parts of the isoleucine biosynthetic pathway, finding that AHAS II (ilvGM) from Salmonella typhimurium and threonine deaminase (ilvA) from Corynebacterium glutamicum improve 2MB production the most. Overexpression of the native threonine biosynthetic operon (thrABC) on plasmid without the native transcription regulation also improved 2MB production in E. coli. Finally, we knocked out competing pathways upstream of threonine production (ΔmetA, Δtdh) to increase its availability for further improvement of 2MB production. This work led to a strain of E. coli that produces 1.25 g/L 2MB in 24 h, a total alcohol content of 3 g/L, and with yields of up to 0.17 g 2MB/g glucose.
Similar content being viewed by others
Introduction
As the global demand for energy continues to rise, biofuels derived from renewable sources have become increasingly important. Currently, most of the emphasis in biofuels is on bio-ethanol as there is a long history for its production (Chandrakant and Bisaria 1998; Galbe and Zacchi 2002; Jarboe et al. 2007). However, increasing attention is being paid to higher alcohols (Miller et al. 1981; Atsumi et al. 2007, 2008) as the potential for a direct replacement of gasoline. Those alcohols have four to five carbons and possess a host of qualities making them more suitable as a liquid fuel than ethanol, including lower vapor pressure, lower hygroscopicity, and higher energy density.
Since the gasoline we use today consists of a finely tuned mixture of components, it is likely that the next-generation fuel to replace gasoline will also be a mixture and the five carbon alcohol 2-methyl-1-butanol (2MB), also known as active amyl alcohol, is likely to be an important component. Previously, 2MB has been considered a minor byproduct found in beer, wine, and other fermented beverages. This is due to the presence of several pyruvate decarboxylase (PDC) enzymes found in the yeast, Saccharomyces cerevisiae, used to ferment these alcoholic beverages, some of which are able to catalyze the decarboxylation of 2-ketoacids larger than pyruvate, especially those 2-ketoacids involved in branched-chain amino acid synthesis (Dickinson et al. 1997, 1998, 2000). PDC converts these 2-ketoacids into aldehydes, which are then reduced to alcohols in one further step using an alcohol dehydrogenase (ADH) enzyme. Until recently (Atsumi et al. 2008), the utility of these higher alcohols as biofuels has not been fully realized. No report has demonstrated the overproduction of 2MB from glucose in a microorganism.
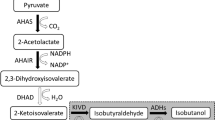
The production of higher alcohols from amino acid precursors in Escherichia coli has proven to be promising (Atsumi et al. 2008; Shen and Liao 2008; Connor and Liao 2008). Because it exploits the host’s native amino acid synthesis pathways, this approach can be implemented in a variety of hosts to produce the desired products. The biosynthetic pathways of the branched-chain amino acids produce at least four natural 2-ketoacids that can be converted to potentially useful higher alcohols (Atsumi et al. 2008). Here, we utilize the isoleucine biosynthetic pathway in particular to synthesize 2-keto-3-methylvalerate (KMV), which is then converted to 2MB via a promiscuous 2-ketoacid decarboxylase (KDC) and ADH (Fig. 1).
Pathway for 2-methyl-1-butanol synthesis and important side products. Genes in bold are overexpressed in our system while underlined genes are knocked out. PEP phosphoenolpyruvate, KIV keto-isovalerate, OAA oxaloacetic acid, αKG α-keto-glutarate, 2KB 2-ketobutyrate, AHB 2-aceto-2-hydroxybutyrate, KMV 2-keto-3-methylvalerate
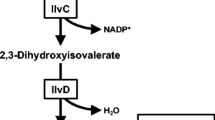
In E. coli, KMV is derived from threonine, which in turn is part of the aspartate family of amino acids. The production of KMV from glucose involves ten enzymatic reactions from the glycolytic product phosphoenolpyruvate (PEP). From PEP, aspartate is produced from the products of two genes, ppc and aspC (Fig. 1). Only four genes, asd and the thrABC operon, are required to complete the five reactions to produce threonine from aspartate. The deamination of threonine to 2-ketobutyrate (2KB) can be catalyzed by the products of either one of two genes, ilvA and tdcB, encoding a biosynthetic and catabolic threonine deaminase, respectively. The condensation of 2KB and pyruvate to 2-aceto-2-hydroxybutyrate is catalyzed by acetohydroxy acid synthase (AHAS), an enzyme which also catalyzes the first step in valine production. Two more genes, ilvC and ilvD, are necessary for the formation of KMV, the penultimate precursor in 2MB synthesis. In the cell, KMV is normally converted to isoleucine with transamination carried out by transaminase B encoded by ilvE.
With the utilization of the isoleucine production pathway in the formation of 2MB, there is a lot of previous work to draw upon on the overproduction of isoleucine in various organisms. It has been shown that overexpression of ilvD is important to prevent the buildup of 2,3-dihydroxy-3-methylvalerate (Hashiguchi et al. 1999), the immediate precursor to KMV. For the condensation of 2KB and pyruvate, there are multiple AHAS enzymes to consider, three in E. coli alone, but each has slightly different properties making it more conducive for either valine or isoleucine production, of which we of course prefer the latter. Of the three isozymes found in E. coli, AHAS II encoded by ilvGM would likely be the best for isoleucine production were it not for a frameshift mutation rendering it inactive in K-12-derived strains (Favre et al. 1976). Nonetheless, enzyme kinetic data show that AHAS II from E. coli and several other enteric bacteria provides a greater ratio of 2-aceto-2-hydroxybutryate to 2-acetolactate formation than the other AHAS isozymes (Barak et al. 1987; Gollop et al. 1990), suggesting that AHAS II should be the best isozyme for isoleucine production. In research on Corynebacterium glutamicum, it was shown that a lysine-producing strain could be converted to isoleucine production with the overexpression of only three key genes: a feedback resistant hom (equivalent to thrA in E. coli), thrB, and feedback resistant ilvA (Morbach et al. 1996). Overexpressing the first gene, thrA, draws carbon flux away from lysine production (Fig. 1), while overexpression of thrB draws carbon flux away from methionine. The third gene, ilvA, allows for the deamination of threonine to 2KB, thus feeding the AHAS enzymatic reaction.
On the basis of the prior art, this article reports the construction of an E. coli strain to produce 2MB from glucose. This work presents additional challenges since byproducts such as 1-propanol and isobutanol are also produced from the KDC and ADH used to produce 2MB. Furthermore, these products are toxic to the cell when present in sufficient concentrations.
Materials and methods
Bacterial strains
E. coli BW25113 (rrnB T14 ΔlacZWJ16 hsdR514 ΔaraBAD AH33 ΔrhaBAD LD78) was designated as the wild-type strain (Datsenko and Wanner 2000) for this study. XL-1 Blue (Stratagene, La Jolla, CA, USA) was used to propagate all plasmids. Construction of strains CRS22 and CRS24 is described previously (Shen and Liao 2008). Strain CRS38 was constructed by deleting the leuABCD operon as previously described (Datsenko and Wanner 2000). Host gene deletions of leuA and ilvE in strains AFC16 and AFC18, respectively, were achieved with P1 transduction using the appropriate Keio collection strains (Baba et al. 2006) as donors. The KanR inserted into the target gene region was removed using pCP20 (Datsenko and Wanner 2000) in between each consecutive knockout. The removal of each gene segment was verified by colony polymerase chain reaction (PCR) using the appropriate primers.
Reagents, media, and cultivation
All restriction enzymes and Antarctic phosphatase were purchased from New England Biolabs (Ipswich, MA, USA). The Rapid DNA ligation kit was supplied by Roche (Manheim, Germany). KOD DNA polymerase was purchased from EMD Chemicals (San Diego, CA, USA). Oligonucleotides were ordered from Invitrogen (Carlsbad, CA, USA).
For all production experiments, strains were grown in a modified M9 medium (6 g Na2HPO4, 3 g KH2PO4, 1 g NH4Cl, 0.5 g NaCl, 1 mM MgSO4, 1 mM CaCl2, 10 mg vitamin B1 per liter of water) containing 15 g/L of glucose, 5 g/L of yeast extract, and 1,000× Trace Metals Mix A5 (2.86 g H3BO3, 1.81 g MnCl2·4H2O, 0.222 g ZnSO4·7H2O, 0.39 g Na2MoO4·2H2O, 79 mg CuSO4·5H2O, 49.4 mg Co(NO3)2·6H2O per liter of water). From 2 mL overnight cultures grown in LB, cultures were inoculated at 1% into 10 mL of the above modified M9 medium and were allowed to grow at 37°C in 250 mL baffled flasks to an OD600 of approximately 0.6 before induction with 0.1 mM IPTG. After induction, 5 mL of the cultures was transferred to 125 mL screw-cap flasks and the incubation temperature was reduced to 30°C. For the experiments where l-threonine or 2-ketobutyrate was added to the culture, it was added at induction to a final concentration of 8 g/L. Antibiotics were added as necessary for plasmid maintenance (ampicillin 100 μg/mL, kanamycin 50 μg/mL, and spectinomycin 50 μg/mL). All antibiotics and reagents were purchased from either Sigma-Aldrich (St. Louis, MO, USA) or Fisher Scientific (Houston, TX, USA).
Plasmid construction
The vectors pSA40, pSA54, pSA55 (Atsumi et al. 2008), and pCS49 (Shen and Liao 2008) were designed and constructed as described previously. The vectors expressing the various AHAS genes were built in a manner similar to pSA54, all beginning with pSA40 as the base.
Vector pKS15 began by amplifying the ilvBN genes from the E. coli MG1655 chromosome using primers IlvB-Acc65I-fwd and IlvNC-SOE1. The beginning of the ilvC gene located on pSA54 was also amplified from the SalI site upstream of the RBS to the Acc65I site within the ilvC gene using primers IlvNC-SOE2 and IlvC-Acc65I-rev. These two fragments were then joined by splice overlap extension (SOE), creating a ilvBNC* fragment with Acc65I cut sites at either end. Then pSA54 was cut with Acc65I, the large portion was gel purified, and then this backbone was ligated with the new ilvBNC* fragment to create pKS15.
In order to test the expression of the protein encoded by E. coli ilvG, we needed to fix the frameshift present at base pair 984 in the ilvG gene found in E. coli MG1655. The sequence at the frameshift indicates a TGA stop codon within the ilvG reading frame, and this sequence was modified from 5′-AATCAATGACTGGCA-3′ to 5′-AATCAATGTGACTGGCA-3′ to recover expression of this protein. To accomplish this feat, we amplified two fragments of the genome with primers IlvG-Acc65I-fwd and TGins-SapI-rev for the beginning portion of the ilvG gene and primers TGins-SapI-fwd and IlvM-HindIII-rev for the end portion of ilvGM. These fragments along with pSA40 were digested with Acc65I, HindIII, and SapI as appropriate and then ligated together to create pAFC22. To complete vector pAFC23, the backbone of pSA54 was amplified from the ilvC gene to the p15A origin of replication using primers IlvC-SA-BamHI-fwd and P15-SA-SacI-rev. This backbone was ligated to the SacI–BamHI fragment from pAFC22 containing the kanamycin resistance gene and ilvGM genes to create pAFC23.
Vectors pAFC3 and pAFC29 each began by using primers IlvG-Acc65I-fwd and IlvM-HindIII-rev to amplify the ilvGM genes from the chromosomes of Salmonella typhimurium LT2 and Klebsiella pneumoniae ATCC 700721, respectively, creating an Acc65I–HindIII fragment. These fragments were then ligated in the same cut sites of pSA40. From here we added the ilvCD genes and P15A replication origin from pSA54 in the same fashion was used for pAFC23.
Vector pAFC26 began by using primers CG-ilvB-ClaI-fwd and CG-ilvN-BamHI-rev to amplify the ilvBN genes from the chromosome of C. glutamicum ATCC 13032 to make a ClaI–BamHI fragment. Ligation of this fragment into the same cut sites of pSA40 introduced a new RBS downstream of the one already present. Similar to the previous three vectors, the ilvCD genes and P15A origin were added to complete this vector.
The vectors expressing the various threonine deaminase genes all used pSA55 as a base. From the E. coli MG1655 chromosome, tdcB (pAFC8) and ilvA (pAFC45) were amplified using primers TdcB-XbaI-fwd/rev and IlvA-XbaI-fwd/rev, respectively. From the C. glutamicum chromosome ilvA (pAFC46) was amplified using primers CG-ilvA-XbaI-fwd/rev. Amplification of each of these three genes resulted in DNA fragments with XbaI cut sites at each end. These fragments were then ligated into the XbaI cut site directly behind the ADH2 gene on pSA55 and orientation was confirmed by PCR and by restriction digest.
All vectors were sequenced using the appropriate primers to verify sequence fidelity.
Metabolite detection
The produced alcohol compounds were quantified by a gas chromatograph (GC) equipped with flame ionization detector (FID). The system consisted of model 5890A GC (Hewlett Packard, Avondale, PA, USA) and a model 7673A automatic injector, sampler and controller (Hewlett Packard). The separation of alcohol compounds was carried out by A DB-WAX capillary column (30 m, 0.32 mm-i.d., 0.50 μm film thickness) purchased from Agilent Technologies (Santa Clara, CA, USA). GC oven temperature was initially held at 40°C for 2 min and raised with a gradient of 5°C/min for 1 min, then held at 45°C for 4 min, then raised again with a gradient of 15°C/min until 190°C. At this point, the temperature was raised with a gradient 50°C/min until 230°C and held for 4 min to clear the column of any remaining chemicals. Helium was used as the carrier gas with 9.0 psi inlet pressure. The FID was fed by a mixture of hydrogen, air, and helium. The injector and detector were maintained at 225°C. The column was injected with 0.5 μL of the supernatant of culture broth in a split injection mode with a 1:15 split ratio. The internal standard used was 1-pentanol, and alcohol content was determined by extrapolation from standard curves using the internal standard to normalize the values.
Results
Selection of AHAS
Our lab has previously shown 2MB formation from glucose in E. coli (Atsumi et al. 2008). In order to increase 2MB production, it was important to first overexpress the enzymes that would boost its production. The first committed reaction in 2MB synthesis is the condensation of 2KB and pyruvate to form 2-aceto-2-hydroxybutyrate. As this reaction can be catalyzed by several different types of AHAS enzyme, we compared a number of these AHAS (Table 1) to select one most suitable for 2MB production.
To test the efficacy of these AHAS isozymes, we replaced ilvIH from pSA54 with other genes encoding AHAS (Table 2) in a cassette fashion to ensure that each enzyme would be expressed similarly. We combined these AHAS plasmids with pSA55, which expresses the KDC and ADH necessary to convert KMV from the isoleucine pathway into 2MB, in a BW25113 host. Since the KDC expressed from pSA55 is able to decarboxylate a broad range of 2-ketoacids (de la Plaza et al. 2004; Atsumi et al. 2008), the KDC and AHAS would compete for 2KB in the cell. If KDC outcompetes AHAS for 2KB, 1-propanol will be produced instead of 2MB. To test the selectivity, we added 0.8% w/v 2KB to the production medium and measured 2MB and 1-propanol production.
The AHAS II from S. typhimurium produced the highest amount of 2MB and is the only AHAS that led to higher selectivity of 2MB production versus 1-propanol. AHAS III from E. coli also produced a significant amount of 2MB, but could not outcompete KDC for 2KB, thus 1-propanol production was even higher. While AHAS I is used in isoleucine production in C. glutamicum, this isozyme did not fare well in 2MB production in E. coli, though it performed much better than the AHAS I from E. coli. The conversion rate of 2KB to 2MB is very low for E. coli AHAS I compared to the other isozymes. Since this enzyme is a poor consumer of 2KB, the high 2KB concentration became poisonous to the cells (LaRossa and Van Dyk 1987). Not only does S. typhimurium AHAS II outcompete the KDC for 2KB, but it also outperforms other competing enzymes for 2KB consumption, making this enzyme the clear favorite among those tested here.
Selection of threonine deaminase
After the AHAS enzyme, the next important enzyme target for overexpression is the threonine deaminase. As shown previously, threonine deaminase overexpression effectively draws carbon flux towards the isoleucine pathway (Morbach et al. 1996). In E. coli, there exists both a biosynthetic (encoded by ilvA) and a catabolic (encoded by tdcB) threonine deaminase. The biosynthetic deaminase is susceptible to feedback inhibition by isoleucine (Umbarger 1956), while the catabolic deaminase is not, though it can be inhibited by high 2KB concentrations (Shizuta et al. 1973). We also compared ilvA from C. glutamicum, though it, too, is inhibited by isoleucine (Möckel et al. 1992).
In a similar fashion as the AHAS comparison, to test these threonine deaminases we inserted each of the three genes into pSA55 in a cassette fashion. As a control we used pSA55, which contains no threonine deaminase gene, and compared this to the effect of the overexpression of threonine deaminase. We then combined these plasmids with pAFC3, containing the genes encoding S. typhimurium AHAS II as determined from the previous experiment, in the same BW25113 host background. We added 0.8% w/vl-threonine to the production medium to study the efficiency of l-threonine deamination by the various isozymes and measured the alcohol production profiles in each case (Fig. 2).
Comparison of threonine deaminase efficiency in 2MB production. Wild-type cells expressing pAFC3 and either pSA55 (no deaminase overexpression), pAFC8 (E. coli tdcB), pAFC45 (E. coli ilvA), or pAFC46 (C. glutamicum ilvA) were supplied with 8 g/L l-threonine. Samples were taken 16 h after induction. Conversion is given as molar conversion from the supplied l-threonine. Data shown are the average of at least three independent experiments
Overexpression of each of the threonine deaminase isozymes results in a greater conversion of l-threonine to 2MB and 1-propanol compared to chromosomal deaminase activity. This is in agreement with previous data suggesting greater isoleucine production with overexpression of threonine deaminase. The increased expression of E. coli biosynthetic deaminase (ilvA) only increased conversion from about 38% to about 45%, while increasing expression of the catabolic deaminase from E. coli (tdcB) converted 62% of the supplied l-threonine. However, the best isozyme tested was the C. glutamicum threonine deaminase, the overexpression of which converted nearly 88% of the supplied l-threonine to 2MB and 1-propanol.
Overproduction of the threonine pathway
One further part of the pathway that would be important to amplify is the formation of threonine from OAA. This transformation occurs in six enzymatic reactions, but previous results (Morbach et al. 1996) indicate that the steps governed by thrA and thrB are the most important. The products of these two genes lie at important branch points in the aspartate family of amino acids (Fig. 1), with thrA drawing carbon flux away from lysine production and thrB drawing carbon flux away from methionine. The thrA gene product also catalyzes the first step in the conversion of aspartate to isoleucine. In E. coli, thrABC is situated in one operon and so we cloned this operon into a low copy number vector in pCS49, removing the native transcription attenuation. Combined with the plasmids determined in the earlier experiments, pAFC46 (KDC, ADH, threonine deaminase) and pAFC3 (AHAS II, ilvCD), we performed 16-h production experiments comparing the effect of adding this threonine operon on 2MB production in (Fig. 3).
In wild-type background with only chromosomal expression of thrABC, isobutanol is the major product, as there is not enough 2KB being produced for sufficient 2MB production. Addition of the threonine operon plasmid doubles the 2MB production from 5.5 to 11 mM, while nearly cutting isobutanol production in half from 21 to 11.5 mM. The formation of 1-propanol also triples from 1.9 to 5.8 mM, which though in itself is not desirable, shows that much more 2KB is being generated in the cultures. In fact, the decrease in isobutanol (9.5 mM) is almost entirely accounted for in the increase of both 2MB and 1-propanol (9.4 mM), indicating that the effect of thrABC overexpression is to pull carbon flux away from pyruvate and towards 2KB.
Gene deletion to attenuate product specificity
Upon determining a useful set of enzymes for overexpression to boost 2MB production, we looked to gene knockouts to further increase 2MB production and yield while reducing side product formation. The first two knockouts upstream of 2KB production, ΔmetA, for elimination of carbon flux to methionine production, which diverts the flux away from threonine, and Δtdh, for reduction of competing threonine consumption, are directed at improving the carbon flux towards 2KB. From this double knockout strain we took two paths towards reducing the alcohol side products made. The first path involved removing the competing native AHAS isozymes that contribute to isobutanol formation, and these are encoded by ilvI and ilvB. The second approach began with eliminating the two peripheral side products, 1-butanol and 3-methyl-1-butanol, by deletion of leuA, which encodes the enzyme catalyzing the first committed step in the formation of these two alcohols. This strain was further engineered by removing ilvE in the hopes of reducing the amount of isoleucine created from the KMV produced. The full results of these gene knockouts are shown in Fig. 4. Each strain shown in Fig. 4 contains the three plasmids (pAFC3, pAFC46, pCS49) expressing all the genes that we determined were beneficial to 2MB production.
Effect of gene knockout combinations on 2-methyl-1-butanol production. Wild-type cells and strains CRS22 (ΔmetA Δtdh), CRS24 (ΔmetA Δtdh ΔilvBI), CRS38 (ΔmetA Δtdh ΔilvBI ΔleuABCD), AFC16 (ΔmetA Δtdh ΔleuA), and AFC18 (ΔmetA Δtdh ΔleuA ΔilvE) expressing pAFC3, pAFC46, and pCS49 are shown. Data shown are the average of at least three independent experiments. a 2MB production. b Isobutanol production. c 1-Propanol production. d Glucose consumption. e Cell density
The strain with the highest 2MB production is the ΔmetA Δtdh double knockout strain, CRS22. It seems that the deletion of just these two genes causes a large increase in 2KB produced as measured by the production of 2MB and 1-propanol. Using this metric, nearly 24 mM 2KB is converted to 2MB and 1-propanol in CRS22 as compared to only 14.5 mM in wild-type. However, much of that extra 2KB goes towards 1-propanol production as 2MB increases from 11.2 to 14.2 mM while 1-propanol increases from 3.2 to 9.6 mM.
As for additional gene deletions, the most noticeable difference is in the diverse alcohol production profiles. From strain CRS22, further deletion of ilvB and ilvI to make CRS24 eliminates the competing AHAS enzymes that contribute more to valine production than to isoleucine. While isobutanol production is reduced by one third, 2MB production is also slightly reduced by about 15% by the loss of these additional AHAS enzymes. However, CRS24 converts the same amount of total 2KB (24 mM) compared to CRS22, as the 1-propanol production rises by exactly the amount of lost 2MB. The leuABCD operon was then removed from CRS24 to make CRS38 in order to completely remove 1-butanol and 3-methyl-1-butanol production. While CRS38 produces a better ratio of 2MB to isobutanol and 1-propanol than the other strains tested, it also suffers from a lower overall production compared to the other strains with a much slower growth rate, and so is not clearly better than the others.
The other string of knockout strains began by deleting leuA directly from the double knockout CRS22 (ΔmetA Δtdh) to make AFC16 for a cleaner product profile and reduce consumption of 2KB for 1-butanol production. Unfortunately, the major effect noticed was lower total cell density compared with the parent strain. While less severe than CRS38, it seems that the deletion of leuA has some adverse effect on growth when combined with the plasmids used in our system. Deletion of leuA also removed all traces of 1-butanol and 3-methyl-1-butanol, but again this apparently did little for the production of the other alcohols. Despite these considerations, this strain converted about the same amount of 2KB (23 mM) as its parent CRS22 and also CRS24, suggesting that increasing the 2KB levels further is more important for producing more 2MB than knocking out these side product pathways. Continuing from AFC16, we next made AFC18 by removing ilvE, which encodes the branched-chain amino acid aminotransferase, looking to increase 2MB production by reducing the amount of isoleucine created from the KMV produced. Strangely, this deletion actually reduced 2MB somewhat but made little difference in 1-propanol and isobutanol production. It was thought that this knockout would particularly benefit 2MB due to the low K M value that this aminotransferase has for KMV (Lee-Peng et al. 1979), but this was not the case. The leuA and ilvE knockouts do not seem to provide much benefit to 2MB production, contrary to what we thought would be the case.
To take a closer look at the production profile of 2MB and related alcohol byproducts, a more detailed time course was obtained of the wt strain and CRS22 (ΔmetA Δtdh), both containing production plasmids pAFC3, pAFC46, and pCS49 (Fig. 5). From this figure, it becomes more obvious that production of 2MB is closely tied to the growth phase. Formation of 1-propanol develops as the 2MB production slows, and this negatively impacts 2MB yields.
Discussion
We successfully engineered the first strain of E. coli to produce the fuel alcohol 2MB to a final level of 1.25 g/L in 24 h and a total alcohol content of over 3 g/L with a yield of up to 0.17 g 2MB per gram glucose, which is 44% of the maximum theoretical yield. Much of the success of producing 2MB is due to the use of the host’s native isoleucine biosynthetic pathway to synthesize KMV, the 2-ketoacid precursor of both isoleucine and 2MB. Earlier work in producing isoleucine from glucose in E. coli has produced yields of up to 0.25 g/g (Hashiguchi et al. 1999), or 0.35 mol/mol. This is similar to what our strains CRS22 (0.32 mol/mol), CRS24 (0.35 mol/mol), and AFC16 (0.34 mol/mol) achieve during the growth phase before 1-propanol production becomes prominent. It turns out that both isoleucine and 2MB share a maximum theoretical yield of 0.8 mol/mol glucose, so we can compare these molar yields head to head.
While our efforts in producing 2MB were aided by previous work in isoleucine production due to an overlap in their biosynthetic pathways, it is important to recognize the unique challenges involved in 2MB production. The first challenge comes from the bifurcated pathway from glucose to 2MB, and the need for the two precursors, pyruvate and 2KB, to be balanced for efficient 2MB production (Fig. 1). Furthermore, both of these necessary precursors lead directly to an unavoidable alcohol side product, with 2KB leading to 1-propanol formation and pyruvate leading to isobutanol formation. Additionally, each of these alcohols is toxic to the host even at fairly low levels, with 2MB in particular affecting E. coli growth rate at concentrations as low as 1 g/L (data not shown).
To illustrate the difference in the biosynthesis of isoleucine and 2MB, the data in Fig. 2 suggest that feedback inhibition due to isoleucine is not an issue for 2MB production. In fact, ilvA from C. glutamicum produced the best in terms of 2MB despite its inhibition by isoleucine. It has also been reported that in isoleucine production, tdcB from E. coli can produce as much as ten-fold better than ilvA from C. glutamicum in a C. glutamicum strain (Guillouet et al. 1999), and we clearly did not see that effect here. This information serves to highlight one aspect of how much 2MB production differs from isoleucine production.
The ultimate goal is to produce higher alcohol biofuels such as 2MB directly from plant biomass, including cellulose and hemicellulose. First, the production of 2MB must be improved by reducing the alcohol side products, especially the 1-propanol production that becomes prominent after the growth phase. At first glance, it seems this is just another obstacle for metabolic engineering to overcome; however, 1-propanol and isobutanol are formed using the KDC and ADH necessary for 2MB production, and isobutanol production is also aided by overexpression of the AHAS enzyme. From this, it becomes apparent that simple metabolic engineering strategy can do only so much to reduce these side products. Since this becomes an issue of substrate competition with both AHAS and KDC, a protein engineering approach appears necessary.
In this first attempt at production of 2MB from a renewable feedstock, the main challenge addressed was that of precursor balancing. Due to the central role of pyruvate in glucose metabolism, we focused instead on increasing the availability of 2KB for efficient 2MB biosynthesis. This was accomplished by overexpressing key genes in the threonine biosynthetic pathway (thrABC) without native transcription regulation and also overexpressing an efficient threonine deaminase to convert the increased threonine to 2KB. Further improvement was obtained by knocking out the competing pathways upstream of 2KB production (ΔmetA, Δtdh). Combined with the overexpression of the downstream part of isoleucine biosynthesis (ilvGMCD), along with the KDC and ADH necessary for conversion of KMV to 2MB, this strategy worked to achieve 1.25 g/L 2MB produced in 24 h.
References
Atsumi S, Cann AF, Connor MR, Shen RC, Simth KM, Brynildsen MP, Chou KC, Hanai T, Liao JC (2007) Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng. doi: https://doi.org/10.1016/j.ymben.2007.08.003
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi:https://doi.org/10.1038/msb4100050
Barak Z, Chipman DM, Gollop N (1987) Physiological implications of the specificity of acetohydroxy acid synthase isozymes of enteric bacteria. J Bacteriol 169:3750–3756
Chandrakant P, Bisaria VS (1998) Simultaneous bioconversion of cellulose and hemicellulose to ethanol. Crit Rev Biotechnol 18:295–331
Connor MR, Liao JC (2008) Engineering Escherichia coli for the production of 3-methyl-1-butanol. Appl Environ Microbiol. doi:https://doi.org/10.1128/AEM.00468-08
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645
de la Plaza M, Fernandez de Palencia P, Pelaez C, Requena T (2004) Biochemical and molecular characterization of a-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol Lett 238:367–374
Dickinson JR, Lanterman MM, Danner DJ, Pearson BM, Sanz P, Harrison SJ, Hewlins MJ (1997) A 13C nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J Biol Chem 272:26871–26878
Dickinson JR, Harrison SJ, Hewlins MJ (1998) An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae. J Biol Chem 273:25751–25756
Dickinson JR, Harrison SJ, Dickinson JA, Hewlins MJ (2000) An investigation of the metabolism of isoleucine to active amyl alcohol in Saccharomyces cerevisiae. J Biol Chem 275:10937–10942
Favre R, Wiater A, Puppo S, Iaccarino M (1976) Expression of a valine-resistant acetolactate synthase activity mediated by the ilv O and ilv G genes of Escherichia coli K-12. Mol Gen Genet 143:243–252
Galbe M, Zacchi G (2002) A review of the production of ethanol from softwood. Appl Microbiol Biotechnol 59:618–628
Gollop N, Damri B, Chipman DM, Barak Z (1990) Physiological implications of the substrate specificities of acetohydroxy acid synthases from varied organisms. J Bacteriol 172:3444–3449
Guillouet S, Rodal AA, An G, Lessard PA, Sinskey AJ (1999) Expression of the Escherichia coli catabolic threonine dehydratase in Corynebacterium glutamicum and its effect on isoleucine production. Appl Environ Microbiol 65:3100–3107
Hashiguchi K, Takesada H, Suzuki E, Matsui H (1999) Construction of an l-isoleucine overproducing strain of Escherichia coli K-12. Biosci Biotechnol Biochem 63:672–679
Jarboe LR, Grabar TB, Yomano LP, Shanmugan KT, Ingram LO (2007) Development of ethanologenic bacteria. Adv Biochem Eng Biotechnol 108:237–261
LaRossa RA, Van Dyk TK (1987) Metabolic mayhem caused by 2-ketoacid imbalances. Bioessays 7:125–130
Lee-Peng FC, Hermodson MA, Kohlhaw GB (1979) Transaminase B from Escherichia coli: quaternary structure, amino-terminal sequence, substrate specificity, and absence of a separate valine-alpha-ketoglutarate activity. J Bacteriol 139:339–345
Miller GL, Smith JL, Workman JP (1981) Engine performance using butanol fuel blends. Trans ASAE 24:538–540
Möckel B, Eggeling L, Sahm H (1992) Functional and structural analyses of threonine dehydratase from Corynebacterium glutamicum. J Bacteriol 174:8065–8072
Morbach S, Sahm H, Eggeling L (1996) l-Isoleucine production with Corynebacterium glutamicum: further flux increase and limitation of export. Appl Environ Microbiol 62:4345–4351
Shen CR, Liao JC (2008) Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab Eng. doi:https://doi.org/10.1016/j.ymben.2008.08.001
Shizuta Y, Kurosawa A, Inoue K, Tanabe T, Hayaishi O (1973) Regulation of biodegradative threonine deaminase. I. Allosteric inhibition of the enzyme by a reaction product and its reversal by adenosin 5'-monophosphate. J Biol Chem 248:512–520
Umbarger HE (1956) Evidence for a negative-feedback mechanism in the biosynthesis of isoleucine. Science 123:848
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Cann, A.F., Liao, J.C. Production of 2-methyl-1-butanol in engineered Escherichia coli . Appl Microbiol Biotechnol 81, 89–98 (2008). https://doi.org/10.1007/s00253-008-1631-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1631-y