Abstract
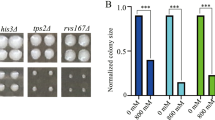
To construct yeast strains showing tolerance to high salt concentration stress, we analyzed the transcriptional response to high NaCl concentration stress in the yeast Saccharomycescerevisiae using DNA microarray and compared between two yeast strains, a laboratory strain and a brewing one, which is known as a stress-tolerant strain. Gene expression dynamically changed following the addition of NaCl in both yeast strains, but the degree of change in the gene expression level in the laboratory strain was larger than that in the brewing strain. The response of gene expression to the low NaCl concentration stress was faster than that to the high NaCl concentration stress in both strains. Expressions of the genes encoding enzymes involved in carbohydrate metabolism and energy production in both strains or amino acid metabolism in the brewing strain were increased under high NaCl concentration conditions. Moreover, the genes encoding sodium ion efflux pump and copper metallothionein proteins were more highly expressed in the brewing strain than in the laboratory strain. According to the results of transcriptome analysis, candidate genes for the creation of stress-tolerant strain were selected, and the effect of overexpression of candidate genes on the tolerance to high NaCl concentration stress was evaluated. Overexpression of the GPD1 gene encoding glycerol-3-phosphate dehydrogenase, ENA1 encoding sodium ion efflux protein, and CUP1 encoding copper metallothionein conferred high salt stress tolerance to yeast cells, and our selection of candidate genes for the creation of stress-tolerant yeast strains based on the transcriptome data was validated.








Similar content being viewed by others
References
Albertyn J, Hohmann S, Thevelein JM, Prior BA (1994) GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by high-osmolarity glycerol response pathway. Mol Cell Biol 14:4135–4144
Ansell R, Granath K, Hohmann S, Thevelein JM, Adler L (1997) The two isoenzymes for yeast NAD+-dependent glycerol-3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J 16:2179–2187
Attfield PV (1997) Stress tolerance: the key to effective strains of industrial baker’s yeast. Nat Biotechnol 15:1351–1357
Blomberg A (2000) Metabolic surprises in Saccharomyces cerevisiae during adaptation to saline conditions: questions, some answers and a model. FEMS Microbiol Lett 182:1–8
Boorstein WR, Craig EA (1990) Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J Biol Chem 265:18912–18921
Butt TR, Sternberg E, Herd J, Crooke ST (1984) Cloning and expression of a yeast copper metallothionein gene. Gene 27:23–33
Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, Weng S, Botstein D (1998) SGD: Saccharomyces genome database. Nucleic Acids Res 26:73–79
Gietz RD, Wodds RA (2001) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96
Goffeau A, Barell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (1996) Life with 6,000 genes. Science 274:546–567
Haslbeck M, Walke S, Stromer T, Ehrnsperger M, White HE, Chen S, Saibil HR, Buchner J (1999) Hsp26: a temperature-regulated chaperone. EMBO J 18:6744–6751
Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66:300–372
Köhrer K, Domdey H (1991) Preparation of high molecular weight RNA. Methods Enzymol 194:398–405
Lewis JG, Learmonth RP, Watson K (1995) Induction of heat, freezing and salt tolerance by heat and salt shock in Saccharomyces cerevisiae. Microbiology 141:687–694
Liu XD, Thiele DJ (1996) Oxidative stress induced heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev 10:592–603
Morita Y, Nakamori S, Takagi H (2002) Effect of proline and arginine metabolism on freezing stress of Saccharomyces cerevisiae. J Biosci Bioeng 94:390–394
Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Ariño J (2000) The transcriptional response of yeast to saline stress. J Biol Chem 275:17249–17255
Rep M, Krantz M, Thevelein JM, Hohmann S (2000) The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem 275:8290–8300
Takagi H, Iwamoto F, Nakamori S (1997) Isolation of freeze-tolerant laboratory strains of Saccharomyces cerevisiae from proline-analogue-resistant mutants. Appl Microbiol Biotechnol 47:405–411
Takagi H, Sakai K, Morida K, Nakamori S (2000) Proline accumulation by mutation or disruption of the proline oxidase gene improves resistance to freezing and desiccation stresses in Saccharomyces cerevisiae. FEMS Microbiol Lett 184:103–108
Varela JCS, Praekelt UM, Meacock PA, Planta RJ, Mager WH (1995) The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol Cell Biol 15:6232–6245
Wieland J, Nitsche AM, Strayle J, Steiner H, Rudolph HK (1995) The PMR gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J 14:3870–3882
Winston F, Dollard C, Ricupero-Hovasse SL (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53–55
Wotton D, Freedman K, Shore D (1996) Multimerization of Hsp42p, a novel heat shock protein of Saccharomyces cerevisiae, is dependent on a conserved carboxyl-terminal sequence. J Biol Chem 271:2717–2723
Yale J, Bohnert HJ (2001) Transcript expression in Saccharomyces cerevisiae at high salinity. J Biol Chem 276:15996–16007
Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J, Lee NH, Yeatman TJ, Quackenbush J (2002) Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol 3:research0062.1–0062.12
Zhao S, Douglas NW, Heine MJ, Williams GM, Winther-Larsen HC, Meaden PG (1994) The STL1 gene of Saccharomyces cerevisiae is predicted to encode a sugar transporter-like protein. Gene 146:215–219
Acknowledgements
This work was carried out as a project of the Development of a Technological Infrastructure for Industrial Bioprocesses of the New Energy and Industrial Technology Development Organization. This work was supported in part by a grant from the 21st century center of excellence program of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hirasawa, T., Nakakura, Y., Yoshikawa, K. et al. Comparative analysis of transcriptional responses to saline stress in the laboratory and brewing strains of Saccharomyces cerevisiae with DNA microarray. Appl Microbiol Biotechnol 70, 346–357 (2006). https://doi.org/10.1007/s00253-005-0192-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-005-0192-6