Abstract
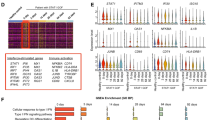
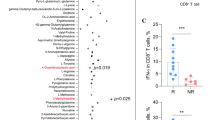
Memory programming of cytotoxic T cells (CTLs) by inflammatory cytokines can be regulated by mammalian target of rapamycin (mTOR). We have shown that inhibition of mTOR during CTL activation leads to the enhancement of memory, but the molecular mechanisms remain largely unknown. Using high-throughput RNA-Seq, we identified genes and functions in mouse CTLs affected by mTOR inhibition through rapamycin. Of the 43,221 identified transcripts, 184 transcripts were differentially expressed after rapamycin treatment, corresponding to 128 annotated genes. Of these genes, 114 were downregulated and only 14 were upregulated. Most importantly, 50 of them are directly related to cell death and survival. In addition, several genes such as CD62L are related to migration. Furthermore, we predicted downregulation of transcriptional regulators based on the total differentially expressed genes, as well as the subset of apoptosis-related genes. Quantitative PCR confirmed the differential expressions detected in RNA-Seq. We conclude that the regulatory function of rapamycin may work through inhibition of multiple genes related to apoptosis and migration, which enhance CTL survival into memory.





Similar content being viewed by others
References
Agarwal P, Raghavan A, Nandiwada SL, Curtsinger JM, Bohjanen PR, Mueller DL, Mescher MF (2009) Gene regulation and chromatin remodeling by IL-12 and type I IFN in programming for CD8 T cell effector function and memory. J Immunol 183:1695–1704
Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R (2009) mTOR regulates memory CD8 T-cell differentiation. Nature 460:108–112
Araki K, Ellebedy AH, Ahmed R (2011) TOR in the immune system. Curr Opin Cell Biol 23:707–715
Badovinac VP, Porter BB, Harty JT (2004) CD8+ T cell contraction is controlled by early inflammation. Nat Immunol 5:809–817
Barbi J, Pardoll D, Pan F (2013) Metabolic control of the Treg/Th17 axis. Immunol Rev 252:52–77
Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, Bourque C, Burke CJ, Turner L, Uong A, Johnson LA, Beroukhim R, Mermel CH, Loda M, Ait-Si-Ali S, Garraway LA, Young RA, Zon LI (2011) The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471:513–517
Chi H (2012) Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol 12:325–338
Curtsinger JM, Lins DC, Mescher MF (2003) Signal 3 determines tolerance versus full activation of naive CD8 T cells: dissociating proliferation and development of effector function. J Exp Med 197:1141–1151
Dejean AS, Hedrick SM, Kerdiles YM (2010) Highly specialized role of Forkhead box O transcription factors in the immune system. Antioxid Redox Signal 14:663–674
Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G (2010) p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol 2:a004887
Dunker N, Schmitt K, Schuster N, Krieglstein K (2002) The role of transforming growth factor beta-2, beta-3 in mediating apoptosis in the murine intestinal mucosa. Gastroenterology 122:1364–1375
Garritano S, Inga A, Gemignani F, Landi S (2013) More targets, more pathways and more clues for mutant p53. Oncogenesis 2:e54
Gillings AS, Balmanno K, Wiggins CM, Johnson M, Cook SJ (2009) Apoptosis and autophagy: BIM as a mediator of tumour cell death in response to oncogene-targeted therapeutics. FEBS J 276:6050–6062
Graziano V, De Laurenzi V (2011) Role of p63 in cancer development. Biochim Biophys Acta 1816:57–66
Hand TW, Cui W, Jung YW, Sefik E, Joshi NS, Chandele A, Liu Y, Kaech SM (2010) Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival. Proc Natl Acad Sci U S A 107:16601–16606
Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177–189
Huang X, Yokota T, Iwata J, Chai Y (2011) Tgf-beta-mediated FasL-Fas-Caspase pathway is crucial during palatogenesis. J Dent Res 90:981–987
Jameson SC, Masopust D (2009) Diversity in T cell memory: an embarrassment of riches. Immunity 31:859–871
Kaech SM, Wherry EJ, Ahmed R (2002) Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol 2:251–262
Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R (2003) Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 4:1191–1198
Kallio JP, Hopkins-Donaldson S, Baker AH, Kahari VM (2011) TIMP-3 promotes apoptosis in nonadherent small cell lung carcinoma cells lacking functional death receptor pathway. Int J Cancer 128:991–996
Kastenmuller W, Brandes M, Wang Z, Herz J, Egen JG, Germain RN (2013) Peripheral prepositioning and local CXCL9 chemokine-mediated guidance orchestrate rapid memory CD8+ T cell responses in the lymph node. Immunity 38:502–513
Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM (2009) Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol 10:176–184
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Li X, Garcia K, Sun Z, Xiao Z (2011) Temporal regulation of rapamycin on memory CTL programming by IL-12. PLoS ONE 6:e25177
Liu N, Phillips T, Zhang M, Wang Y, Opferman JT, Shah R, Ashton-Rickardt PG (2004) Serine protease inhibitor 2A is a protective factor for memory T cell development. Nat Immunol 5:919–926
Liu M, Guo S, Hibbert JM, Jain V, Singh N, Wilson NO, Stiles JK (2011) CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev 22:121–130
Lohr K, Moritz C, Contente A, Dobbelstein M (2003) p21/CDKN1A mediates negative regulation of transcription by p53. J Biol Chem 278:32507–32516
Majumder S, Bhattacharjee S, Paul Chowdhury B, Majumdar S (2012) CXCL10 is critical for the generation of protective CD8 T cell response induced by antigen pulsed CpG-ODN activated dendritic cells. PLoS ONE 7:e48727
Menke J, Iwata Y, Rabacal WA, Basu R, Yeung YG, Humphreys BD, Wada T, Schwarting A, Stanley ER, Kelley VR (2009) CSF-1 signals directly to renal tubular epithelial cells to mediate repair in mice. J Clin Invest 119:2330–2342
Mescher MF, Curtsinger JM, Agarwal P, Casey KA, Gerner M, Hammerbeck CD, Popescu F, Xiao Z (2006) Signals required for programming effector and memory development by CD8+ T cells. Immunol Rev 211:81–92
Michaud JL, Rosenquist T, May NR, Fan CM (1998) Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev 12:3264–3275
Obar JJ, Lefrancois L (2010) Early signals during CD8(+) T cell priming regulate the generation of central memory cells. J Immunol 185:263–272
Ozsolak F, Milos PM (2011) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12:87–98
Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y (2009) Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460:103–107
Rao RR, Li Q, Odunsi K, Shrikant PA (2010) The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity 32:67–78
Singh UP, Singh S, Singh R, Cong Y, Taub DD, Lillard JW Jr (2008) CXCL10-producing mucosal CD4+ T cells, NK cells, and NKT cells are associated with chronic colitis in IL-10(−/−) mice, which can be abrogated by anti-CXCL10 antibody inhibition. J Interferon Cytokine Res 28:31–43
Staal FJ, Luis TC, Tiemessen MM (2008) WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol 8:581–593
Sung JH, Zhang H, Moseman EA, Alvarez D, Iannacone M, Henrickson SE, de la Torre JC, Groom JR, Luster AD, von Andrian UH (2012) Chemokine guidance of central memory T cells is critical for antiviral recall responses in lymph nodes. Cell 150:1249–1263
Thomson AW, Turnquist HR, Raimondi G (2009) Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol 9:324–337
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53
Watford WT, Hissong BD, Bream JH, Kanno Y, Muul L, O’Shea JJ (2004) Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunol Rev 202:139–156
Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R (2003) Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol 4:225–234
Williams MA, Bevan MJ (2007) Effector and memory CTL differentiation. Annu Rev Immunol 25:171–192
Xiang H, Wang J, Boxer LM (2006) Role of the cyclic AMP response element in the bcl-2 promoter in the regulation of endogenous Bcl-2 expression and apoptosis in murine B cells. Mol Cell Biol 26:8599–8606
Xiao Z, Casey KA, Jameson SC, Curtsinger JM, Mescher MF (2009) Programming for CD8 T cell memory development requires IL-12 or type I IFN. J Immunol 182:2786–2794
Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H (2013) mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499:485–490
Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH (2010) Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 33:229–240
Acknowledgments
We thank Mr. Ken Class for technical assistance. This work was partially supported by the National Institutes of Health GrantsR21AI095715A (to X. Z.) and Startup from UMD (to X. Z.). This work was supported in part by AFRI grant No. 2011-67015-30183 from USDA NIFA (to GEL). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. The USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Elliot Mattson and Lingyang Xu are co-first authors.
Rights and permissions
About this article
Cite this article
Mattson, E., Xu, L., Li, L. et al. Transcriptome profiling of CTLs regulated by rapamycin using RNA-Seq. Immunogenetics 66, 625–633 (2014). https://doi.org/10.1007/s00251-014-0790-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-014-0790-5