Abstract
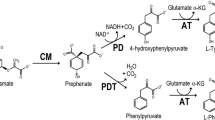
Phenylalanine hydroxylase (PAH) is a non-heme iron enzyme that catalyzes oxidation of phenylalanine to tyrosine, a reaction that must be kept under tight regulatory control. Mammalian PAH has a regulatory domain in which binding of the substrate leads to allosteric activation of the enzyme. However, the existence of PAH regulation in evolutionarily distant organisms, for example some bacteria in which it occurs, has so far been underappreciated. In an attempt to crystallographically characterize substrate binding by PAH from Chromobacterium violaceum, a single-domain monomeric enzyme, electron density for phenylalanine was observed at a distal site 15.7 Å from the active site. Isothermal titration calorimetry (ITC) experiments revealed a dissociation constant of 24 ± 1.1 μM for phenylalanine. Under the same conditions, ITC revealed no detectable binding for alanine, tyrosine, or isoleucine, indicating the distal site may be selective for phenylalanine. Point mutations of amino acid residues in the distal site that contact phenylalanine (F258A, Y155A, T254A) led to impaired binding, consistent with the presence of distal site binding in solution. Although kinetic analysis revealed that the distal site mutants suffer discernible loss of their catalytic activity, X-ray crystallographic analysis of Y155A and F258A, the two mutants with the most noticeable decrease in activity, revealed no discernible change in the structure of their active sites, suggesting that the effect of distal binding may result from protein dynamics in solution.









Similar content being viewed by others
Abbreviations
- PAH:
-
Phenylalanine hydroxylase
- BH4 :
-
(6R)-5,6,7,8-tetrahydrobiopterin dihydrochloride
- PKU:
-
Phenylketonuria
- cPAH:
-
Phenylalanine hydroxylase from Chromobacterium violaceum
- hPAH:
-
Human phenylalanine hydroxylase
- RMSD:
-
Root-mean-square deviation
- THA:
-
3-(2-thienyl)-l-alanine
- NLE:
-
l-Norleucine
- Fe:
-
Iron
- Co:
-
Cobalt
- DMPH4 :
-
6,7-Dimethyltetrahydropterin
References
Abu-Omar MM, Loaiza A, Hontzeas N (2005) Reaction mechanisms of mononuclear non-heme iron oxygenases. Chem Rev 105:2227–2252
Adler-Abramovich L, Vaks L, Carny O, Trudler D, Magno A, Caflisch A, Frenkel D, Gazit E (2012) Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat Chem Biol 8:701–706
Andersen OA, Flatmark T, Hough E (2002) Crystal structure of the ternary complex of the catalytic domain of human phenylalanine hydroxylase with tetrahydrobiopterin and 3-(2-thienyl)-l-alanine, and its implications for the mechanism of catalysis and substrate activation. J Mol Biol 320:1095–1108
Andersen OA, Stokka AJ, Flatmark T, Hough E (2003) 2.0 Ǻ resolution crystal structures of the ternary complexes of human phenylalanine hydroxylase catalytic domain with tetrahydrobiopterin and 3-(2-Thienyl)-l-alanine or l-Norleucine: substrate specificity and molecular motions related to substrate binding. J Mol Biol 333:747–757
Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem 97:10269–10280
Broecker J, Vargas C, Keller S (2011) Revisiting the optimal c value for isothermal titration calorimetry. Anal Biochem 418:307–309
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang RE et al (2012) AMBER12. University of California, San Francisco
Chehin R, Thorolfsson M, Knappskog PM, Martinez A, Flatmark T, Arrondo JL, Muga A (1998) Domain structure and stability of human phenylalanine hydroxylase inferred from infrared spectroscopy. FEBS Lett 422:225–230
Citron BA, Davis MD, Kaufman S (1992) Purification and biochemical characterization of recombinant rat liver phenylalanine hydroxylase produced in Escherichia coli. Protein Exp Purif 3:93–100
Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763
Cooper A, Dryden DTF (1984) Allostery without conformational change: a plausible model. Eur Biophys J 11:103–109
Daubner SC, Hillas PJ, Fitzpatrick P (1997) Characterization of chimeric pterin-dependent hydroxylases: contributions of the regulatory domains of tyrosine and phenylalanine hydroxylase to substrate specificity. Biochemistry 36:11574–115782
DeLano WL (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501
Erlandsen H, Kim JY, Patch MG, Han A, Volner A, Abu-Omar MM, Stevens RC (2002) Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates. J Mol Biol 320:645–661
Erlandsen H, Patch MG, Gamez A, Straub M, Stevens RC (2003) Structural studies on phenylalanine hydroxylase and implications toward understanding and treating phenylketonuria. Pediatrics 112:1557–1565
Fitzpatrick PF (1999) Tetrahydropterin-dependent amino acid hydroxylases. Annu Rev Biochem 68:355–381
Fitzpatrick PF (2003) Mechanism of aromatic amino acid hydroxylation. Biochemistry 42:14083–14091
Fitzpatrick PF (2012) Allosteric regulation of phenylalanine hydroxylase. Arch Biochem Biophys 519:194–201
Flatmark T, Stevens RC (1999) Structural insight into the aromatic amino acid hydroxylases and their disease-related mutant forms. Chem Rev 99:2137–2160
Flydal MI, Mohn TC, Pey AL, Siltberg-Liberles J, Teigen K, Martinez A (2010) Superstoichiometric binding of l-Phe to phenylalanine hydroxylase from Caenorhabditis elegans: evolutionary implications. Amino Acids 39:1463–1475
Fuchs JE, Huber RG, von Grufenstein S, Wallnoefer HG, Spitzer GM, Fuchs D, Liedl KR (2012) Dynamic regulation of phenylalanine hydroxylase by simulated redox manipulation. PLoS ONE 7(12):e53005
Fusetti F, Erlandsen H, Flatmark T, Stevens RC (1998) Structure of tetrameric human phenylalanine hydroxylase and its implications for phenylketonuria. J Biol Chem 273:16962–16967
Gjetting T, Petersen M, Guldberg P, Guttler F (2001) Missense mutations in the N-terminal domain of phenylalanine hydroxylase interfere with binding of regulatory phenylalanine. Am J Hum Genet 68:1353–1360
Goetz AW, Williamson MJ, Xu D, Poole D, Le Grand S et al (2012) Routine microsecond molecular dynamics with AMBER—Part I: generalized Born. J Chem Theory Comput 8(5):1542–1555
Hardy JA, Wells JA (2004) Searching for new allosteric sites in enzymes. Curr Opin Struct Biol 14:706–715
Herrmann KM (1995) The Shikimate pathway: early steps in the biosynthesis of aromatic compounds. Plant Cell 7:907–919
Horne J, Jennings IG, Teh T, Gooley PR, Kobe B (2002) Structural characterization of the N-terminal autoregulatory sequence of phenylalanine hydroxylase. Protein Sci 11:2041–2047
Houtman JC, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE, Schuck P (2007) Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci 16:30–42
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Kappock TJ, Cardonna JP (1996) Pterin-dependent amino acid hydroxylases. Chem Rev 96:2659–2756
Kaufman S (1993) The phenylalanine hydroxylating system. Adv Enzymol Relat Areas Mol Biol 67:77–264
Knappskog PM, Flatmark T, Aarden JM, Haavik J, Martinez A (1996) Structure/function relationships in human phenylalanine hydroxylase. Effect of terminal deletions on the oligomerization, activation and cooperativity of substrate binding to the enzyme. Eur J Biochem 242:813–821
Kobe B, Jennings IG, House CM, Michell BJ, Goodwill KE, Santarsiero BD, Stevens RC, Cotton RGH, Kemp BE (1999) Structural basis of autoregulation of phenylalanine hydroxylase. Nat Struct Biol 6:442–448
Labute P (2009) Protonate 3D: assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins 75(1):187–205
Leiros H-KS, Pey AL, Innselset M, Moe E, Leiros I, Steen IH, Martinez A (2007) Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. J Biol Chem 282:21973–21986
Li J, Ilangovan U, Daubner SC, Hinck AP, Fitzpatrick PF (2011) Direct evidence for a phenylalanine site in the regulatory domain of phenylalanine hydroxylase. Arch Biochem Biophys 505:250–255
Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Dror RO et al (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78(8):1950–1958
Loaiza A, Armstrong KM, Baker BM, Abu-Omar MM (2008) Kinetics of thermal unfolding of phenylalanine hydroxylase variants containing different metal cofactors (Fe(II), Co(II), and Zn(II)) and their isokinetic relationship. Inorg Chem 47:4877–4883
Mayers JR, Fyfe I, Schuh AL, Chapman ER, Edwardson JM, Audhya A (2011) ESCRT-0 assembles as a heterotetrameric complex on membranes and binds multiple ubiquitinylated cargoes simultaneously. J Biol Chem 286:9636–9645
Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367
Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM (eds) Methods in enzymology, macromolecular crystallography, part A, vol 276. Academic Press, New York
Painter J, Merritt EA (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Cryst D62:439–450
Parniak MA, Kaufman S (1981) Rat liver phenylalanine hydroxylase. Activation by sulfhydryl modification. J Biol Chem 256:6876–6882
Phillips RS, Parniak MA, Kaufman S (1984) The interaction of aromatic amino acids with rat liver phenylalanine hydroxylase. J Biol Chem 259:271–277
Scriver CR (1995) Whatever happened to PKU? Clin Biochem 28:137–144
Singh B (1999) In: Singh K (ed) Plant amino acids: biochemistry and biotechnology, Marcel Dekker, Inc, New York
Tsai CJ, Sol AD, Nussinov R (2008) Allostery: absence of a change in shape does not imply that allostery is not at play. J Mol Biol 378: 1–11
Tzeng SR, Kalodimos CG (2011) Protein dynamics and allostery: an NMR view. Curr Opin Struct Biol 21:62–67
Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30:1022–1025
Vasconcelos AT, Almeida DF, Hungria M, Guimarães CT, Antônio RV et al (2003) The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc Natl Acad Sci USA 100:11660–11665
Volner A, Zoidakis J, Abu-Omar MM (2003) Order of substrate binding in bacterial phenylalanine hydroxylase and its mechanistic implication for pterin-dependent oxygenases. J Biol Inorg Chem 8:121–128
Wallnoefer HG, Handschuh S, Liedl KR, Fox T (2010) Stabilizing of a globular protein by a highly complex water network: a molecular dynamics study on factor Xa. J Phys Chem B 114:7405–7412
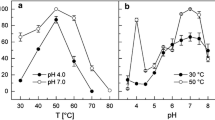
Zoidakis J, Loaiza A, Vu K, Abu-Omar MM (2005) Effect of temperature, pH, and metals on the stability and activity of phenylalanine hydroxylase from Chromobacterium violaceum. J Inorg Biochem 99(3):771–775
Acknowledgments
We are grateful to our hosts Michael Becker, Craig Ogata, Nukri Sanishvili, and Nagarajan Venugopalan at the GM/CA CAT beamline 23-ID-D at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under contract no. DE-AC02-06CH11357. Julian E. Fuchs is a recipient of a DOC fellowship of the Austrian Academy of Sciences at the University of Innsbruck. The authors acknowledge the High Performance Computing at the University of Innsbruck for providing computer hardware resources for this project. This research was funded by the National Science Foundation (CHE-0749572 to M.M.A.O.) and the National Institutes of Health (1R01RR026273 to C.D.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ronau, J.A., Paul, L.N., Fuchs, J.E. et al. An additional substrate binding site in a bacterial phenylalanine hydroxylase. Eur Biophys J 42, 691–708 (2013). https://doi.org/10.1007/s00249-013-0919-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-013-0919-8