Abstract
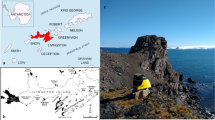
We assessed fungal diversity present in air samples obtained from King George Island, Antarctica, using DNA metabarcoding through high-throughput sequencing. We detected 186 fungal amplicon sequence variants (ASVs) dominated by the phyla Ascomycota, Basidiomycota, Mortierellomycota, Mucoromycota, and Chytridiomycota. Fungi sp. 1, Agaricomycetes sp. 1, Mortierella parvispora, Mortierella sp. 2, Penicillium sp., Pseudogymnoascus roseus, Microdochium lycopodinum, Mortierella gamsii, Arrhenia sp., Cladosporium sp., Mortierella fimbricystis, Moniliella pollinis, Omphalina sp., Mortierella antarctica, and Pseudogymnoascus appendiculatus were the most dominant ASVs. In addition, several ASVs could only be identified at higher taxonomic levels and may represent previously unknown fungi and/or new records for Antarctica. The fungi detected in the air displayed high indices of diversity, richness, and dominance. The airborne fungal diversity included saprophytic, mutualistic, and plant and animal opportunistic pathogenic taxa. The diversity of taxa detected reinforces the hypothesis that the Antarctic airspora includes fungal propagules of both intra- and inter-continental origin. If regional Antarctic environmental conditions ameliorate further in concert with climate warming, these fungi might be able to reactivate and colonize different Antarctic ecosystems, with as yet unknown consequences for ecosystem function in Antarctica. Further aeromycological studies are necessary to understand how and from where these fungi arrive and move within Antarctica and if environmental changes will encourage the development of non-native fungal species in Antarctica.




Similar content being viewed by others
References
Siegert MJ et al (2008) Recent advances in understanding Antarctic climate evolution. Antarct Sci 4:313–325
Convey P (2017) Antarctic ecosystems. In: Reference module in life sciences. Elsevier. https://doi.org/10.1016/B978-0-12-809633-8.02182-8
Archer SDJ, Lee KC, Caruso T et al (2018) Microbial dispersal limitation to isolated soil habitats in the McMurdo Dry Valleys of Antarctica. bioRxiv 493411. https://doi.org/10.1101/493411
Marshall WA (1996) Aerial dispersal of lichen soredia in the maritime Antarctic. New Phytol 134:523–530
Vincent WF (2000) Evolutionary origins of Antarctic microbiota: invasion, selection and endemism. Antarct Sci 12:374–385
de Menezes GCA, Porto BA, Simões JC, Rosa CA, Rosa LH (2019a) Fungi in snow and glacial ice of Antarctica. In: Rosa LH (ed) Fungi of Antarctica1st edn. Springer, Basel, pp 127–146
de Menezes GCA, Porto BA, Amorim SS, Zani CL, de Almeida Alves TM, Junior PAS, Murta SMF, Simões JC, Cota BB, Rosa CA, Rosa LH (2020) Fungi in glacial ice of Antarctica: diversity, distribution and bioprospecting of bioactive compounds. Extremophiles 24:367–376
Rosa LH et al (2020a) DNA metabarcoding of fungal diversity in air and snow of Livingston Island, South Shetland Islands, Antarctica. Sci. Rep. in press
Sundberg S (2013) Spore rain in relation to regional sources and beyond. Ecography 36:364–373
Bottos EM, Woo AC, Zawar-Reza P, Pointing SB, Cary SC (2014) Airborne bacterial populations above desert soils of the McMurdo dry valleys, Antarctica. Microb Ecol 67:120–128
Pearce DA et al (2016) Aerobiology over Antarctica - a new initiative for atmospheric ecology. Front Microbiol 7:16
Ruisi S, Barreca D, Selbmann L, Zucconi L, Onofri S (2007) Fungi in Antarctica. Rev Environ Sci Biotechnol 6:127–141
Rosa LH, Zani CL, Cantrell CL, Duke SO, van Dijck P, Desideri A, Rosa CA (2019) Fungi in Antarctica: diversity, ecology, effects of climate change, and bioprospection for bioactive compounds. In: Rosa LH (ed) Fungi of Antarctica: diversity, Ecology and Biotechnological Applications. Springer, Cham, pp 1–18
Marshall WA (1997) Seasonality in Antarctic airborne fungal spores. Appl Environ Microbiol 63:2240–2245
Hughes KA, McCartney H, Lachlan-Cope TA, Pearce DA (2004) A preliminary study of airborne microbial biodiversity over peninsular Antarctica. Cell Mol Biol 50:537–542
Pearce DA, Hughes KA, Lachlan-Cope T, Harangozo SA, Jones AE (2010) Biodiversity of airborne microorganisms at Halley Station, Antarctica. Extremophiles 14:145–159
Turner J, Lu H, White I, King JC, Phillips T, Hosking JS, Bracegirdle TJ, Marshall GJ, Mulvaney R, Deb P (2016) Absence of 21st century warming on Antarctic peninsula consistent with natural variability. Nature 535:411–415
Bracegirdle TJ, Connolley WM, Turner J (2008) Antarctic climate change over the twenty first century. J Geophys Res 113:D03103
Convey P, Peck LS (2019) Antarctic environmental change and biological responses. Sci Adv 5:eaaz0888
Siegert M et al (2019) The Antarctic peninsula under a 1.5°C global warming scenario. Front. Environ Sci 7:102
Chen S, Yao H, Han J, Liu C, Song J, Shi L, Zhu Y, Ma X, Gao T, Pang X, Luo K, Li Y, Li X, Jia X, Lin Y, Leon C (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5:e8613
Richardson RT, Lin CH, Sponsler DB, Quijia JO, Goodell K, Johnson RM (2015) Application of ITS2 metabarcoding to determine the provenance of pollen collected by honey bees in an agroecosystem. Appl Plant Sci 3:1400066
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Joshi NA, Fass JN (2011) Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software]. https://github.com/najoshi/sickle
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson II MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277
Medinger R et al (2010) Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol 19:32–40
Giner CR, Forn I, Romac S, Logares R, de Vargas C, Massana R (2016) Environmental sequencing provides reasonable estimates of the relative abundance of specific picoeukaryotes. Appl Environ Microbiol 82:4757–4766
Weber AA, Pawlowski J (2013) Can abundance of protists be inferred from sequence data: a case study of Foraminifera. PLoS One 8:e56739
Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière-Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, Vere N, Pfrender ME, Bernatchez L (2017) Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol Ecol 26:5872–5895
Hering D, Borja A, Jones JI, Pont D, Boets P, Bouchez A, Bruce K, Drakare S, Hänfling B, Kahlert M, Leese F, Meissner K, Mergen P, Reyjol Y, Segurado P, Vogler A, Kelly M (2018) Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Res 138:192–205
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:9
Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C (2014) An interactive Venn diagram viewer. BMC Bioinformatics 15:293
Rosa LH et al (2020b) DNA metabarcoding uncovers fungal diversity in soils of protected and non-protected areas on Deception Island, Antarctica. Sci. Rep. in press
Duncan SM, Farrell RL, Jordan N, Jurgens JA, Blanchette RA (2010) Monitoring and identification of airborne fungi at historic locations on Ross Island, Antarctica. Polar Sci 4:275–283
Seifert KA et al (2011) The genera of Hyphomycetes. CBS-KNAW Fungal Biodiversity Centre, Utrecht, p 997
Mahuku GS, Hsiang T, Yang L (1998) Genetic diversity of Microdochium nivale isolates from turfgrass. Mycol Res 102:559–567
Hernández-Restrepo M, Groenewald JZ, Crous PW (2016) Taxonomic and phylogenetic re-evaluation of Microdochium, Monographella and Idriella. Persoonia 36:57–82
Gonçalves VN, Vaz ABM, Rosa CA, Rosa LH (2012) Diversity and distribution of fungal communities in lakes of Antarctica. FEMS Microbiol Ecol 82:459–471
Santiago IF, Alves TMA, Rabello A, Sales Junior PA, Romanha AJ, Zani CL, Rosa CA, Rosa LH (2012) Leishmanicidal and antitumoral activities of endophytic fungi associated with the Antarctic angiosperms Deschampsia antarctica Desv. and Colobanthus quitensis (Kunth) Bartl. Extremophiles 16:95–103
Walter J, Hermann V (2012) Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales). Fungal Divers 52:75–98
Carvalho CR et al (2020) Fungi associated with the briosphere of the bipolar mosses Polytrichastrum alpinum and Polytrichum juniperinum in Antarctica. Polar Biol 43:545–553
Mercantini R, Marsella R, Cervellati MC (1989) Keratinophilic fungi isolated from Antarctic soil. Mycopathologia 106:47–52
Lorch JM, Lindner DL, Gargas A, Muller LK, Minnis AM, Blehert DS (2013) A culture-based survey of fungi in soil from bat hibernacula in the eastern United States and its implications for detection of Geomyces destructans, the causal agent of bat white-nose syndrome. Mycologia 105:237–252
Minnis AM, Lindner DL (2013) Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biol 117:638–649
Arenz BE, Blanchette RA (2011) Distribution and abundance of soil fungi in Antarctica at sites on the Peninsula, Ross Sea Region and McMurdo Dry Valleys. Soil Biol Biochem 43:308–315
Krishnan A, Alias SA, Wong CMVL, Pang KL, Convey P (2011) Extracellular hydrolase enzyme production by soil fungi from King George Island, Antarctica. Polar Biol 34:1535–1542
Gomes EC et al (2018) Cultivable fungi present in Antarctic soils: taxonomy, phylogeny, diversity, and bioprospecting of antiparasitic and herbicidal metabolites. Extremophiles 22:381–393
Tosi S, Casado B, Gerdol R (2002) Fungi isolated from Antarctic mosses. Polar Biol 25:262–268
Rosa LH, Almeida Vieira Mde L, Santiago IF, Rosa CA (2010) Endophytic fungi community associated with the dicotyledonous plant Colobanthus quitensis (Kunth) Bartl. (Caryophyllaceae) in Antarctica. FEMS Microbiol Ecol 73:178–189
Loque CP, Medeiros AO, Pellizzari FM, Oliveira EC, Rosa CA, Rosa LH (2010) Fungal community associated with marine macroalgae from Antarctica. Polar Biol 33:641–648
Furbino LE, Godinho VM, Santiago IF, Pellizari FM, Alves TMA, Zani CL, Junior PAS, Romanha AJ, Carvalho AGO, Gil LHVG, Rosa CA, Minnis AM, Rosa LH (2014) Diversity patterns, ecology and biological activities of fungal communities associated with the endemic macroalgae across the Antarctic. Microb Ecol 67:775–787
Santiago IF, Soares MA, Rosa CA, Rosa LH (2015) Lichenosphere: a protected natural microhabitat of the non-lichenised fungal communities living in extreme environments of Antarctica. Extremophiles 19:1087–1097
McRae CF, Hocking AD, Seppelt RD (1999) Penicillium species from terrestrial habitats in the Windmill Islands, East Antarctica, including a new species, Penicillium antarcticum. Polar Biol 21:97–111
Godinho VM, Gonçalves VN, Santiago IF, Figueredo HM, Vitoreli GA, Schaefer CEGR, Barbosa EC, Oliveira JG, Alves TMA, Zani CL, Junior PAS, Murta SMF, Romanha AJ, Kroon EG, Cantrell CL, Wedge DE, Duke SO, Ali A, Rosa CA, Rosa LH (2015) Diversity and bioprospection of fungal community present in oligotrophic soil of continental Antarctica. Extremophiles 19:585–596
Zucconi L, Selbmann L, Buzzini P, Turchetti B, Guglielmin M, Frisvad JC, Onofri S (2012) Searching for eukaryotic life preserved in Antarctic permafrost. Polar Biol 35:749–757
Silva TH et al (2020) Diversity, distribution, and ecology of viable fungi in permafrost and active layer of Maritime Antarctica. Extremophiles 24:565–576
Godinho VM, Furbino LE, Santiago IF, Pellizzari FM, Yokoya NS, Pupo D, Alves TMA, S Junior PA, Romanha AJ, Zani CL, Cantrell CL, Rosa CA, Rosa LH (2013) Diversity and bioprospecting of fungal communities associated with endemic and cold-adapted macroalgae in Antarctica. ISME J 7:1434–1451
Acknowledgments
This study received financial support from CNPq, PROANTAR, FAPEMIG, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES), INCT Criosfera 2. P. Convey is supported by NERC core funding to the British Antarctic Survey’s “Biodiversity, Evolution and Adaptation” Team. We also thank congresswoman Jô Moraes and the Biological Sciences Institute of the University of Brasilia.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rosa, L.H., Pinto, O.H.B., Convey, P. et al. DNA Metabarcoding to Assess the Diversity of Airborne Fungi Present over Keller Peninsula, King George Island, Antarctica. Microb Ecol 82, 165–172 (2021). https://doi.org/10.1007/s00248-020-01627-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01627-1