Abstract
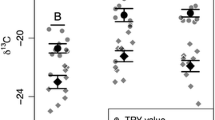
Dryland ecosystems are increasing in geographic extent and contribute greatly to interannual variability in global carbon dynamics. Disentangling interactions among dominant primary producers, including plants and autotrophic microbes, can help partition their contributions to dryland C dynamics. We measured the δ13C signatures of biological soil crust cyanobacteria and dominant plant species (C3 and C4) across a regional scale in the southwestern USA to determine if biocrust cyanobacteria were coupled to plant productivity (using plant-derived C mixotrophically), or independent of plant activity (and therefore purely autotrophic). Cyanobacterial assemblages located next to all C3 plants and one C4 species had consistently more negative δ13C (by 2‰) than the cyanobacteria collected from plant interspaces or adjacent to two C4 Bouteloua grass species. The differences among cyanobacterial assemblages in δ13C could not be explained by cyanobacterial community composition, photosynthetic capacity, or any measured leaf or root characteristics (all slopes not different from zero). Thus, microsite differences in abiotic conditions near plants, rather than biotic interactions, remain a likely mechanism underlying the observed δ13C patterns to be tested experimentally.



Similar content being viewed by others
References
Jackson RB, Lajtha K, Crow SE et al (2017) The ecology of soil carbon: pools, vulnerabilities, and biotic and abiotic controls. Annu Rev Ecol Evol Syst 48:419–445. https://doi.org/10.1146/annurev-ecolsys-112414-054234
Bardgett RD, Wardle DA (2010) Aboveground-belowground linkages: biotic interactions, ecosystem processes, and global change. Oxford University Press, Oxford, New York
Classen AT, Sundqvist MK, Henning JA et al (2015) Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: What lies ahead? Ecosphere 6:130. https://doi.org/10.1890/ES15-00217.1
van der Heijden MGA, Bardgett RD, van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310. https://doi.org/10.1111/j.1461-0248.2007.01139.x
Zhang Z-S, Zhao Y, Dong X-J, Shi YF, Chen YL, Hu YG (2016) Evolution of soil respiration depends on biological soil crusts across a 50-year chronosequence of desert revegetation. Soil Sci Plant Nutr 62:140–149. https://doi.org/10.1080/00380768.2016.1165597
Rudgers JA, Dettweiler-Robinson E, Belnap J, Green LE, Sinsabaugh RL, Young KE, Cort CE, Darrouzet-Nardi A (2018) Are fungal networks key to dryland primary production? Am J Bot 105:1783–1787. https://doi.org/10.1002/ajb2.1184
Sancho LG, Belnap J, Colesie C, Raggio J, Weber B (2016) Carbon budgets of biological soil crusts at micro-, meso-, and global scales. In: Weber B, Büdel B, Belnap J (eds) Biological soil crusts: an organizing principle in drylands. Springer International Publishing, Cham, pp 287–304
Ahlström A, Xia J, Arneth A, Luo Y, Smith B (2015) Importance of vegetation dynamics for future terrestrial carbon cycling. Environ Res Lett 10:054019. https://doi.org/10.1088/1748-9326/10/5/054019
Elbert W, Weber B, Burrows S, Steinkamp J, Büdel B, Andreae MO, Pöschl U (2012) Contribution of cryptogamic covers to the global cycles of carbon and nitrogen. Nat Geosci 5:459–462. https://doi.org/10.1038/ngeo1486
Chamizo S, Canton Y, Lazaro R et al (2012) Crust composition and disturbance drive infiltration through biological soil crusts in semiarid ecosystems. Ecosystems 15:148–161. https://doi.org/10.1007/s10021-011-9499-6
Belnap J, Weber B, Büdel B (2016) Biological soil crusts as an organizing principle in drylands. In: Weber B, Büdel B, Belnap J (eds) Biological soil crusts: an organizing principle in drylands. Springer International Publishing, Cham, pp 3–13
Machado de Lima NM, Fernandes VMC, Roush D et al (2019) The compositionally distinct cyanobacterial biocrusts from Brazilian Savanna and their environmental drivers of community diversity. Front Microbiol 10. https://doi.org/10.3389/fmicb.2019.02798
Garcia-Pichel F, Loza V, Marusenko Y, Mateo P, Potrafka RM (2013) Temperature drives the continental-scale distribution of key microbes in topsoil communities. Science 340:1574–1577. https://doi.org/10.1126/science.1236404
Nagy ML, Perez A, Garcia-Pichel F (2005) The prokaryotic diversity of biological soil crusts in the Sonoran Desert (Organ Pipe Cactus National Monument, AZ). FEMS Microbiol Ecol 54:233–245. https://doi.org/10.1016/j.femsec.2005.03.011
Bowker MA, Belnap J, Davidson DW, Harland G (2006) Correlates of biological soil crust abundance across a continuum of spatial scales: support for a hierarchical conceptual model. J Appl Ecol 43:152–163. https://doi.org/10.1111/j.1365-2664.2006.01122.x
Collins SL, Belnap J, Grimm NB, Rudgers JA, Dahm CN, D'Odorico P, Litvak M, Natvig DO, Peters DC, Pockman WT, Sinsabaugh RL, Wolf BO (2014) A multiscale, hierarchical model of pulse dynamics in arid-land ecosystems. Annu Rev Ecol Evol Syst 45:397–419. https://doi.org/10.1146/annurev-ecolsys-120213-091650
Garcia-Pichel F, Belnap J (1996) Microenvironments and microscale productivity of cyanobacterial desert crusts. J Phycol 32:774–782
Green LE, Porras-Alfaro A, Sinsabaugh RL (2008) Translocation of nitrogen and carbon integrates biotic crust and grass production in desert grassland. J Ecol 96:1076–1085. https://doi.org/10.1111/j.1365-2745.2008.01388.x
Garcia-Pichel F (2009) Cyanobacteria. Encyclopedia of Microbiology. Elsevier, pp 107–124. https://www-sciencedirect-com.libproxy.unm.edu/science/article/pii/B9780123739445002509?via%3Dihub
Baran R, Brodie EL, Mayberry-Lewis J, Hummel E, da Rocha UN, Chakraborty R, Bowen BP, Karaoz U, Cadillo-Quiroz H, Garcia-Pichel F, Northen TR (2015) Exometabolite niche partitioning among sympatric soil bacteria. Nat Commun 6:8289. https://doi.org/10.1038/ncomms9289
Couradeau E, Giraldo-Silva A, De Martini F, Garcia-Pichel F (2019) Spatial segregation of the biological soil crust microbiome around its foundational cyanobacterium, Microcoleus vaginatus, and the formation of a nitrogen-fixing cyanosphere. Microbiome 7:55. https://doi.org/10.1186/s40168-019-0661-2
Dawson TE, Mambelli S, Plamboeck AH, Templer PH, Tu KP (2002) Stable isotopes in plant ecology. Annu Rev Ecol Syst 33:507–559. https://doi.org/10.1146/annurev.ecolsys.33.020602.095451
Diefendorf AF, Mueller KE, Wing SL, Koch PL, Freeman KH (2010) Global patterns in leaf 13C discrimination and implications for studies of past and future climate. Proc Natl Acad Sci 107:5738–5743. https://doi.org/10.1073/pnas.0910513107
Ziegler H, Lüttge U (1998) Carbon isotope discrimination in cyanobacteria of rocks of inselbergs and soils of savannas in the neotropics. Bot Acta 111:212–215
Darby BJ, Neher DA (2012) Stable isotope composition of microfauna supports the occurrence of biologically fixed nitrogen from cyanobacteria in desert soil food webs. J Arid Environ 85:76–78. https://doi.org/10.1016/j.jaridenv.2012.06.006
Lange OL, Kilian E, Ziegler H (1986) Water vapor uptake and photosynthesis of lichens: performance differences in species with green and blue-green algae as phycobionts. Oecologia 71:104–110
Dettweiler-Robinson E (2018) Biocrust carbon isotope signature was depleted under a C3 forb compared to interspace. Plant Soil 429:101–111. https://doi.org/10.1007/s11104-017-3558-5
Harris D, Horwáth WR, van Kessel C (2001) Acid fumigation of soils to remove carbonates prior to total organic carbon or CARBON-13 isotopic analysis. Soil Sci Soc Am J 65:1853–1856. https://doi.org/10.2136/sssaj2001.1853
Fernandes VMC, Machado de Lima NM, Roush D, Rudgers J, Collins SL, Garcia-Pichel F (2018) Exposure to predicted precipitation patterns decreases population size and alters community structure of cyanobacteria in biological soil crusts from the Chihuahuan Desert: Changing rainfall effects on soil cyanobacteria. Environ Microbiol 20:259–269. https://doi.org/10.1111/1462-2920.13983
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. https://doi.org/10.1038/ismej.2012.8
Bolyen E, Rideout JR, Dillon MR et al (2018) QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. https://doi.org/10.7287/peerj.preprints.27295v2
DeSantis TZ, Hugenholtz P, Larsen N et al (2006) Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl Environ Microbiol 72:5069–5072. https://doi.org/10.1128/AEM.03006-05
Roush D (2018) Fgplab/Cydrasil: V1.0.1-Zen. Zenodo
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. https://doi.org/10.1093/nar/gkw290
Allen HK, Bayles DO, Looft T, Trachsel J, Bass BE, Alt DP, Bearson SMD, Nicholson T, Casey TA (2016) Pipeline for amplifying and analyzing amplicons of the V1–V3 region of the 16S rRNA gene. BMC Res Notes 9:380. https://doi.org/10.1186/s13104-016-2172-6
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Aust Ecol 26:32–46. https://doi.org/10.1111/j.1442-9993.2001.01070.pp.x
Castle SC, Morrison CD, Barger NN (2011) Extraction of chlorophyll a from biological soil crusts: A comparison of solvents for spectrophotometric determination. Soil Biol Biochem 43:853–856. https://doi.org/10.1016/j.soilbio.2010.11.025
Soundararajan M (2012) Leaf chlorophyll levels influence carbon isotope discrimination in soybean and maize. Int J Biosci Biochem Bioinforma:207–211. https://doi.org/10.7763/IJBBB.2012.V2.102
Yevdokimov IV, Ruser R, Buegger F, Marx M, Munch JC (2007) Carbon turnover in the rhizosphere under continuous plant labeling with 13CO2: partitioning of root, microbial, and rhizomicrobial respiration. Eurasian Soil Sci 40:969–977. https://doi.org/10.1134/S1064229307090074
Yang W, Magid J, Christensen S, Rønn R, Ambus P, Ekelund F (2014) Biological 12C–13C fractionation increases with increasing community-complexity in soil microcosms. Soil Biol Biochem 69:197–201. https://doi.org/10.1016/j.soilbio.2013.10.030
Schlesinger WH, Pilmanis AM (1998) Plant-soil interactions in deserts. Biogeochemistry 42:169–187
Ochoa-Hueso R, Eldridge DJ, Delgado-Baquerizo M, Soliveres S, Bowker MA, Gross N, le Bagousse-Pinguet Y, Quero JL, García-Gómez M, Valencia E, Arredondo T, Beinticinco L, Bran D, Cea A, Coaguila D, Dougill AJ, Espinosa CI, Gaitán J, Guuroh RT, Guzman E, Gutiérrez JR, Hernández RM, Huber-Sannwald E, Jeffries T, Linstädter A, Mau RL, Monerris J, Prina A, Pucheta E, Stavi I, Thomas AD, Zaady E, Singh BK, Maestre FT (2018) Soil fungal abundance and plant functional traits drive fertile island formation in global drylands. J Ecol 106:242–253. https://doi.org/10.1111/1365-2745.12871
Börjesson G, Menichetti L, Thornton B, Campbell CD, Kätterer T (2016) Seasonal dynamics of the soil microbial community: assimilation of old and young carbon sources in a long-term field experiment as revealed by natural 13 C abundance: seasonal changes among microbes in agricultural soil. Eur J Soil Sci 67:79–89. https://doi.org/10.1111/ejss.12309
Liao JD, Boutton TW, Jastrow JD (2006) Organic matter turnover in soil physical fractions following woody plant invasion of grassland: Evidence from natural 13C and 15 N. Soil Biol Biochem 38:3197–3210. https://doi.org/10.1016/j.soilbio.2006.04.004
Bai E, Boutton TW, Liu F, Wu XB, Hallmark CT, Archer SR (2012) Spatial variation of soil δ13C and its relation to carbon input and soil texture in a subtropical lowland woodland. Soil Biol Biochem 44:102–112. https://doi.org/10.1016/j.soilbio.2011.09.013
Kramer C, Gleixner G (2006) Variable use of plant- and soil-derived carbon by microorganisms in agricultural soils. Soil Biol Biochem 38:3267–3278. https://doi.org/10.1016/j.soilbio.2006.04.006
Rippka R, Deruelles J, Waterbury J et al (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1–61
Bernard L, Mougel C, Maron P-A, Nowak V, Lévêque J, Henault C, Haichar FZ, Berge O, Marol C, Balesdent J, Gibiat F, Lemanceau P, Ranjard L (2007) Dynamics and identification of soil microbial populations actively assimilating carbon from 13 C-labelled wheat residue as estimated by DNA- and RNA-SIP techniques. Environ Microbiol 9:752–764. https://doi.org/10.1111/j.1462-2920.2006.01197.x
Lee CG, Watanabe T, Fujita Y, Asakawa S, Kimura M (2012) Heterotrophic growth of cyanobacteria and phage-mediated microbial loop in soil: examination by stable isotope probing (SIP) method. Soil Sci Plant Nutr 58:161–168. https://doi.org/10.1080/00380768.2012.658739
Ehleringer JR (1993) Carbon and water relations in desert plants: an isotopic perspective. Stable Isotopes and Plant Carbon-water Relations. Elsevier, pp 155–172. https://doi.org/10.1016/B978-0-08-091801-3.50018-0
Acknowledgments
We thank Jarek Kwiecinski, Noelle Martinez, Laura Green, Charlie Mettler, Vishwa Patel, Maria Isabel Siles Asaff, for lab and field work assistance. We thank Dr. Benjamin Brunner at UTEP and Laura Burkemper, Seth Newsome, and Viorel Atudorei for stable isotope analysis assistance.
Availability of Data and Material
Data is available from EDI (https://doi.org/10.6073/pasta/b59b733653671fc1a28f12a865e121bc) and sequences are available at NCBI (SRA number SUB5948515).
Funding
Funding was provided by NSF nos. 1557135, 1557162, and Jornada LTER grant nos. 1832194. This research was partially supported by grants from the National Science Foundation to the University of New Mexico for Long-term Ecological Research (SEV-LTER).
Author information
Authors and Affiliations
Contributions
ES, JR, RS, FG-P, JB, and AD-N made substantial contributions to the conception or design of the work; GC, VF, CN, AG-S contributed acquisition, analysis, and interpretation of data; all contributed to drafting the work and revisions.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
Not applicable
Consent to Participate
Not applicable
Consent for Publication
Not applicable
Code Availability
Code is available at https://github.com/evadeeter/cyanobacteria-13C.
Additional information
Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Electronic supplementary material
ESM 1
(DOCX 341 kb)
Rights and permissions
About this article
Cite this article
Stricker, E., Crain, G., Rudgers, J. et al. What Could Explain δ13C Signatures in Biocrust Cyanobacteria of Drylands?. Microb Ecol 81, 134–145 (2021). https://doi.org/10.1007/s00248-020-01536-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01536-3