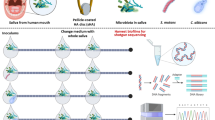
Abstract
Oral diseases are biofilm-mediated diseases caused by imbalances in the ecology of resident microflora. Among them, dental caries (tooth decay) is considered the most common disease worldwide, and toothbrushing, which physically eliminates the oral biofilm, is the most widespread prevention strategy. Although it is well established that fluoride increases enamel resistance to acidic pH and promotes tooth remineralization, its effect on the biofilm bacterial communities’ composition and metabolism is not fully understood. We have grown in vitro oral biofilms and used 16S rRNA Illumina sequencing to study the effect of fluoride on DNA- and RNA-based bacterial populations. In addition, a metatranscriptomic approach has also been performed, in which total RNA has been sequenced to study gene expression profiles in the presence/absence of 500 ppm sodium fluoride. Our data show a lower pH drop and a clear shift in total and metabolically active bacterial composition after fluoride exposure. Streptococcus oralis was the species most affected, with a 10-fold reduction in both DNA and RNA samples, whereas Rothia mucilaginosa underwent an 8-fold increase in the DNA and S. salivarius a 4- and 5-fold increase in the RNA and DNA samples, respectively. The metatranscriptomes indicated that fluoride exposure induced a dramatic shutdown of sugar metabolism, including significant under-expression of different sugar transporters, fucosidases, and a pyruvate oxidase, among others. The reduction in saccharolytic organisms and the inhibition of sugar fermentation pathways by fluoride may therefore be considered instrumental for the beneficial effect of fluoride-containing oral hygiene products.




Similar content being viewed by others
References
Berger D, Rakhamimova A, Pollack A, Loewy Z (2018) Oral biofilms: development, control, and analysis. High-Throughput 7:24. https://doi.org/10.3390/ht7030024
Xu X, Chen F, Huang Z, Ma L, Chen L, Pan Y et al (2018) Meeting report: a close look at oral biofilms and microbiomes. J Oral Sci 10:28. https://doi.org/10.1038/s41368-018-0030-1
Marsh PD (2006) Dental plaque as a biofilm and a microbial community–implications for health and disease. BMC Oral Health 6:S14. https://doi.org/10.1186/1472-6831-6-S1-S14
ten Cate JM (2013) Contemporary perspective on the use of fluoride products in caries prevention. Br Dent J 214:161–167. https://doi.org/10.1038/sj.bdj.2013.162
Marquis RE, Clock SA, Mota-Meira M (2003) Fluoride and organic weak acids as modulators of microbial physiology. FEMS Microbiol Rev 26:493–510. https://doi.org/10.1111/j1574-69762003tb00627x
Van Loveren C (2001) Antimicrobial activity of fluoride and its in vivo importance: identification of research questions. Caries Res 35(Suppl 1):65–70. https://doi.org/10.1159/000049114
Reilly C, Goettl M, Steinmetz M, Nikrad J, Jones RS (2016) Short-term effects of povidone iodine and sodium fluoride therapy on plaque levels and microbiome diversity. Oral Dis 22:155–161. https://doi.org/10.1111/odi12407
Koopman JE et al (2015) The effect of fixed orthodontic appliances and fluoride mouthwash on the oral microbiome of adolescents–a randomized controlled clinical trial. PLoS One 10:e0137318. https://doi.org/10.1371/journalpone0137318
Rosin-Grget K, Peros K, Sutej I, Basic K (2013) The cariostatic mechanisms of fluoride. Acta Med Acad 42:179–188. https://doi.org/10.5644/ama2006-12485
Tian Y et al (2010) Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol Oral Microbiol 25:357–367. https://doi.org/10.1111/j2041-1014201000585x
Ferrer MD et al (2016) Effect of antibiotics on biofilm inhibition and induction measured by real-time cell analysis. J Appl Microbiol 122:640–650. https://doi.org/10.1111/jam13368
Mira A et al (2019) Development of an in vitro system to study oral biofilms in real time through impedance technology: validation and potential applications. J Oral Microbiol 11:1609838. https://doi.org/10.1080/2000229720191609838
Nyvad B, Crielaard W, Mira A, Takahashi N, Beighton D (2013) Dental caries from a molecular microbiological perspective. Caries Res 47:89–102. https://doi.org/10.1159/000345367
Lagerlof F, Oliveby A, Ekstrand J (1987) Physiological factors influencing salivary clearance of sugar and fluoride. J Dent Res 66:430–435. https://doi.org/10.1177/00220345870660020801
Dzidic M et al (2018) Oral microbiome development during childhood: an ecological succession influenced by postnatal factors and associated with tooth decay. ISME J 12:2292–2306. https://doi.org/10.1038/s41396-018-0204-z
Benítez-Páez A, Belda-Ferre P, Simón-Soro A, Mira A (2014) Microbiota diversity and gene expression dynamics in the human oral biofilm. BMC Genomics 15:311–323. https://doi.org/10.1186/1471-2164-15-311
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. https://doi.org/10.1093/bioinformatics/btr026
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM00062-07
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth3869
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124. https://doi.org/10.1093/bioinformatics/btu494
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Quast C et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Langmead B, Salzberg S (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth1923
Li D et al (2016) MEGAHIT v10: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 1:3–11. https://doi.org/10.1016/jymeth201602020
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular biology open software suite. Trends Genet 16:276–277
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. https://doi.org/10.1093/nar/28127
Durbin R, Eddy SR, Krogh A, Mitchison GJ (1998) Biological sequence analysis: probabilistic models of proteins and nucleic acids. UK Cambridge University Press, Cambridge
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Li R et al (2017) Comparison of DNA-, PMA-, and RNA-based 16S rRNA Illumina sequencing for detection of live bacteria in water. Sci Rep 7:5752. https://doi.org/10.1038/s41598-017-02516-3
Naumova EA et al (2012) Fluoride bioavailability in saliva and plaque. BMC Oral Health 12:3. https://doi.org/10.1186/1472-6831-12-3
Rosier BT, Marsh PD, Mira A (2018) Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J Dent Res 97:371–380. https://doi.org/10.1177/0022034517742139
Wescombe PA, Hale JD, Heng NC, Tagg JR (2012) Developing oral probiotics from Streptococcus salivarius. Future Microbiol 7:1355–1371. https://doi.org/10.2217/fmb12113
Liao Y, Brandt BW, Li J, Crielaard W, Van Loveren C, Deng DM (2017) Fluoride resistance in Streptococcus mutans: a mini review. J Oral Microbiol 9(1):1344509
Klug B, Santigli E, Westendorf C et al (2016) From mouth to model: combining in vivo and in vitro oral biofilm growth. Front Microbiol 21:1448
Simón-Soro A, Belda-Ferre P, Cabrera-Rubio R, Alcaraz LD, Mira A (2013) A tissue-dependent hypothesis of dental caries. Caries Res 47:591–600
Hamilton IR (1990) Biochemical effects of fluoride on oral bacteria. J Dent Res 69:660–667; discussion 682-3. https://doi.org/10.1177/00220345900690S128
Takahashi N, Washio J (2011) Metabolomic effects of xylitol and fluoride on plaque biofilm in vivo. J Dent Res 90:1463–1468. https://doi.org/10.1177/0022034511423395
Belli WA, Marquis RE (1994) Catabolite modification of acid tolerance of Streptococcus mutans GS-5. Oral Microbiol Immunol 9:29–34. https://doi.org/10.1111/j1399-302x1994tb00211x
Murata T, Hanada N (2016) Contribution of chloride channel permease to fluoride resistance in Streptococcus mutans. FEMS Microbiol Lett 363:fnw101. https://doi.org/10.1093/femsle/fnw101
Huang X, Schulte RM, Burne RA, Nascimento MM (2015) Characterization of the arginolytic microflora provides insights into pH homeostasis in human oral biofilms. Caries Res 49:165–176. https://doi.org/10.1159/000365296
Burne RA, Marquis RE (2000) Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett 193:1–6. https://doi.org/10.1111/j.1574-6968.2000.tb09393.x
López-López A, Camelo-Castillo A, Ferrer MD, Simón-Soro A, Mira A (2017) Health-associated niche inhabitants as oral probiotics: the case of Streptococcus dentisani. Front Microbiol 8:379. https://doi.org/10.3389/fmicb201700379
Nascimento MM et al (2019) Metabolic profile of supragingival plaque exposed to arginine and fluoride. J Dent Res 98:1245–1252. https://doi.org/10.1177/0022034519869906
Huang X et al (2017) Effect of arginine on the growth and biofilm formation of oral bacteria. Arch Oral Biol 82:256–262. https://doi.org/10.1016/jarchoralbio201706026
Thurnheer T, Belibasakis GN (2018) Effect of sodium fluoride on oral biofilm microbiota and enamel demineralization. Arch Oral Biol 89:77–83. https://doi.org/10.1016/jarchoralbio201802010
Acknowledgments
We especially appreciate the involvement of Núria Jiménez and Javier Pons in the massive sequencing and bioinformatic analyses, respectively.
Funding
The research leading to these results has received funding from the Spanish Government under grant agreement RTI 2018-102032-B-100.
Author information
Authors and Affiliations
Contributions
AM and ALL conceived the work. ALL worked in the designing and performance of the experiments. AM and ALL made the analyses and interpretation of data and wrote the manuscript.
Corresponding author
Ethics declarations
All saliva donors signed an informed consent form and the protocol was approved by the Ethics Committee of the Valencian Public Health Authority (FISABIO-DGSP).
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 106 kb)
Rights and permissions
About this article
Cite this article
López-López, A., Mira, A. Shifts in Composition and Activity of Oral Biofilms After Fluoride Exposure. Microb Ecol 80, 729–738 (2020). https://doi.org/10.1007/s00248-020-01531-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01531-8