Abstract
Uric acid is a promising hydrophobic nitrogen source for biostimulation of microbial activities in oil-impacted marine environments. This study investigated metabolic processes and microbial community changes in a series of microcosms using sediment from the Mediterranean and the Red Sea amended with ammonium and uric acid. Respiration, emulsification, ammonium and protein concentration measurements suggested a rapid production of ammonium from uric acid accompanied by the development of microbial communities containing hydrocarbonoclastic bacteria after 3 weeks of incubation. About 80 % of uric acid was converted to ammonium within the first few days of the experiment. Microbial population dynamics were investigated by Ribosomal Intergenic Spacer Analysis and Illumina sequencing as well as by culture-based techniques. Resulting data indicated that strains related to Halomonas spp. converted uric acid into ammonium, which stimulated growth of microbial consortia dominated by Alcanivorax spp. and Pseudomonas spp. Several strains of Halomonas spp. were isolated on uric acid as the sole carbon source showed location specificity. These results point towards a possible role of halomonads in the conversion of uric acid to ammonium utilized by hydrocarbonoclastic bacteria.




Similar content being viewed by others
References
Anderson DG, McKay LL (1983) Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol 46:549–552
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2005) At least 1 in 20, 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71:7724–7736
Atlas EL (1975) Phosphate equilibria in seawater and interstitial waters. PhD thesis. Oregon State Univ., Corvallis, Oregon.
Atlas RM, Bartha R (1972) Degradation and mineralization of petroleum by two bacteria isolated from coastal waters. Biotechnol Bioeng 14:297–308
Capone DG, Bronk DA, Mulholland MR & Carpenter EJ (2008) Nitrogen in the marine environment. (Academic Press, 2008). at http://www.sciencedirect.com/science/book/9780123725226.
Cardinale M et al (2004) Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl Environ Microbiol 70:6147–6156
Clarke KR, Gorley RN (2006) PRIMER v6: user manual/tutorial. PRIMER-E, Plymouth
Dyksterhouse SE, Gray JP, Herwig RP, Lara JC, Staley JT (1995) Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int J Syst Bacteriol 45:116–123
Eck RV, Dayhoff MO (1966) Atlas of protein sequence and structure. National Biomedical Research Foundation, Silver Springs
Fava F, Berselli S, Conte P, Piccolo A, Marchetti L (2004) Effects of humic substances and soya lecithin on the aerobic bioremediation of a soil historically contaminated by polycyclic aromatic hydrocarbons (PAHs). Biotechnol Bioeng 88:214–223
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gagnon K, Rothäusler E, Syrjänen A, Yli-Renko M, Jormalainen V (2013) Seabird fertilizes Baltic Sea littoral food webs. PLoS ONE 8:e61284
Garcia HE, Locarnini RA, Boyer TP, Antonov JI, Zweng MM, Baranova OK & Johnson DR (2010) World Ocean Atlas 2009, Volume 4: Nutrients (phosphate, nitrate, silicate). S. Levitus, Ed. NOAA Atlas NESDIS 71, U.S. Government Printing Office, Washington, D.C.
Gertler C, Gerdts G, Timmis KN, Yakimov MM, Golyshin PN (2009) Populations of heavy fuel oil-degrading marine microbial community in presence of oil sorbent materials. J Appl Microbiol 107:590–605
Gertler C, Gerdts G, Timmis KN, Golyshin PN (2009) Microbial consortia in mesocosm bioremediation trial using oil sorbents, slow-release fertilizer and bioaugmentation. FEMS Microbiol Ecol 69:288–300
Gertler C, Näther DJ, Cappello S, Gerdts G, Quilliam RS, Yakimov MM, Golyshin PN (2012) Composition and dynamics of biostimulated indigenous oil-degrading microbial consortia from the Irish, North and Mediterranean Seas: a mesocosm study. FEMS Microbiol Ecol 81:520–536
Gutierrez T, Singleton DR, Berry D, Yang T, Aitken MD, Teske A (2013) Hydrocarbon-degrading bacteria enriched by the deepwater horizon oil spill identified by cultivation and DNA-SIP. ISME J 7:2091–2104
Gutierrez T, Berry D, Yang T, Mishamandani S, McKay L, Teske A, Aitken MD (2013) Role of bacterial exopolysaccharides (EPS) in the fate of the oil released during the Deepwater Horizon Oil Spill. PLoS ONE 8:e67717
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: palaeontological statistics software package for education and data analysis. Palaeontol Electron 4:9
Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK et al (2010) Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208
Head IM, Jones DM, Röling WFM (2006) Marine microorganisms make a meal of oil. Nat Rev Microbiol 4:173–182
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:277–280
Kanehisa M (1997) A database for post-genome analysis. Trends Genet 13:375–376
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S et al (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:354–357
Kasai Y, Kishira H, Sasaki T, Syutsubo K, Watanabe K, Harayama S (2002) Predominant growth of Alcanivorax strains in oil-contaminated and nutrient-supplemented sea water. Environ Microbiol 4:141–147
Kleinsteuber S, Riis V, Fetzer I, Harms H, Müller S (2006) Population dynamics within a microbial consortium during growth on diesel fuel in saline environments. Appl Environ Microbiol 72:3531–3542
Knezevich V, Koren O, Ron EZ, Rosenberg E (2006) Petroleum bioremediation in seawater using guano as the fertilizer. Bioremed J 10:83–91
Koren O, Knezevic V, Ron EZ, Rosenberg E (2003) Petroleum pollution bioremediation using water-insoluble uric acid as the nitrogen source. Appl Environ Microbiol 69:6337–6339
Kube M, Chernikova TN, Al-Ramahi Y, Beloqui A, Lopez-Cortez N, Guazzaroni ME, Heipieper HJ, Klages S, Kotsyurbenko OR, Langer I, Nechitaylo TY, Lünsdorf H, Fernández M, Juárez S, Ciordia S, Singer A, Kagan O, Egorova O, Petit PA, StogiosP KY, Tchigvintsev A, Flick R, Denaro R, Genovese M, Albar JP, Reva ON, Martínez-Gomariz M, Tran H, Ferrer M, Savchenko A, Yakunin AF, Yakimov MM, Golyshina OV, Reinhardt R, Golyshin PN (2013) Genome sequence and functional genomics analysis of the oil-degrading bacterium Oleispira antarctica. Nat Commun 4:2156
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 21:2947–2948
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Lee K, Tremblay GH, Levy EM (1993) Bioremediation: application of slow-release fertilizers on low-energy shorelines. Int Oil Spill Conf Proc 1993:449–454
Lindstrom JE et al (1991) Microbial populations and hydrocarbon biodegradation potentials in fertilized shoreline sediments affected by the T/V Exxon Valdez oil spill. Appl Environ Microbiol 57:2514–2522
Mata JA, Martínez-Cánovas J, Quesada E, Béjar VA (2002) Detailed phenotypic characterisation of the type strains of Halomonas species. Syst Appl Microbiol 25:360–375
Miranda KM, Espey MG, Wink DA (2001) A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5:62–71
Mnif S, Chamkha M, Sayadi S (2009) Isolation and characterization of Halomonas sp. strain C2SS100, a hydrocarbon-degrading bacterium under hypersaline conditions. J Appl Microbiol 107:785–794
Mulvaney, R. (1996) in Methods of soil analysis part 3: chemical methods. 1123–1184 Soil Science Society of America, Inc.
Nahm KH (2003) Evaluation of the nitrogen content in poultry manure. World’s Poult Sci J 59:77–88
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Nikolopoulou M, Kalogerakis N (2008) Enhanced bioremediation of crude oil utilizing lipophilic fertilizers combined with biosurfactants and molasses. Mar Pollut Bull 56:1855–1861
Nikolopoulou M, Pasadakis N, Kalogerakis N (2013) Evaluation of autochthonous bioaugmentation and biostimulation during microcosm-simulated oil spills. Mar Pollut Bull 72:165–173
Noguchi H, Park J et al (2006) MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 34:5623–5630
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Tamaki H, Matsuoka T, Yasuda Y, Hanada S, Kamagata Y, Nakamura K, Sakasegawa S (2010) A novel laccase with urate oxidation activity from Lysobacter sp. T-15. J Biochem 148:481–489
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Vogels GD, Van der Drift C (1976) Degradation of purines and pyrimidines by microorganisms. Bacteriol Rev 40:403–468
Wang Z, Hollebone BP, Fingas M, Fieldhouse B, Sigouin L, Landriault M, SmithP, Noonan J & Thouin G (2003) Characteristics of spilled oils, fuels, and petroleum products: 1. Composition and properties of selected oils. United States Environmental Protection Agency, National Exposure Research Laboratory, EPA/600/R-03/072
Wrenn BA, Zheng H, Kohar ES, Lee K, Venosa AD (2001) Effect of pulsed additions of nutrients on oil biodegradation in continuous-flow beach microcosms. Int Oil Spill Conf Proc 2001:339–344
Yakimov MM, Gentile G, Bruni V, Cappello S, D’Auria G, Golyshin PN, Giuliano L (2004) Crude oil-induced structural shift of coastal bacterial communities of Rod Bay (Terra Nova Bay, Ross Sea, Antarctica) and characterization of cultured cold-adapted hydrocarbonoclastic bacteria. FEMS Microbiol Ecol 49:419–432
Yakimov MM, Timmis KN, Golyshin PN (2007) Obligate oil-degrading marine bacteria. Curr Opin Biotechnol 18:257–266
Acknowledgments
The authors would like to thank Anna Foster, Sarah Chesworth and Gordon Turner for their help with photometric and respiration measurements. With exception of XH and JC, all authors were supported by the FP7 Project ULIXES (FP7-KBBE-2010-266473). This work was further funded by grant BIO2011-25012 from the Spanish Ministry of the Economy and Competitiveness. FM was supported by Università degli Studi di Milano, European Social Fund (FSE) and Regione Lombardia (contract “Dote Ricerca”). DD acknowledges support of KAUST, King Abdullah University of Science and Technology. PG acknowledges the support of the European Commission through the project Kill-Spill (FP7, Contract Nr 312139). CG would like to thank Mr. Kyungsun Lee of Macrogen Inc. for his courtesy regarding sequencing services, J. Cans, B. Strid and C. Hudson for continued advice and inspiration as well as Delphine Lallias for her help with multivariate statistics and John Flannery for proofreading this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Table 1a
ANOSIM results of bacterial community RISA fingerprinting profiles on the basis of Bray-Curtis distance measure, showing the univariate partition of factors “Location”, “Nitrogen source” and “Sampling day” and the multivariate partition of factors “Location” within the factor “Nitrogen source”. The R-value was calculated based on the Null hypothesis of no similarity between samples. The values highlighted in bold are statistically significant (P < 0.05). (XLSX 9 kb)
Supplementary Table 1b
R-values derived from ANOSIM pairwise comparisons of factor “(sampling) Location” values using Bray-Curtis values. The R-value was calculated based on the Null hypothesis of no similarity between samples. The values highlighted in bold are statistically significant (P < 0.05). (XLSX 9 kb)
Supplementary Table 1c
R-values derived from ANOSIM pairwise comparisons of factor “Sampling day” values using Bray-Curtis values. The R-value was calculated based on the Null hypothesis of no similarity between samples. The values highlighted in bold are statistically significant (P < 0.05). (XLSX 9 kb)
Supplementary Table 2a
Data statistics for samples as obtained by Illumina sequencing (XLSX 10 kb)
Supplementary Table 2b
Data statistics for the best assembly results (XLSX 10 kb)
Supplementary Table 2c
Gene prediction results (XLSX 9 kb)
Supplementary Table 3a
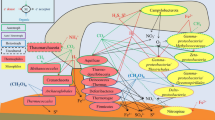
Annotation of the metagenomic data from the Aqaba UA sample from day 21, displayed in Fig. 3 of the main text. The subset consists of 30 genes involved in uric acid metabolism from a total of 26,866 ORFs identified in the metagenome. Gene sequences with similarity to Halomonas spp. are printed in bold. (XLSX 10 kb)
Supplementary Table 3b
Annotation of the metagenomic data from the Ancona UA sample from day 21, displayed in Fig. 3 of the main text. The subset consists of 15 genes involved in uric acid metabolism from a total of 27,893 ORFs identified in the metagenome. Gene sequences with similarity to Halomonas spp. are printed in bold. (XLSX 10 kb)
Supplementary Table 3c
Annotation of the metagenomic data from the Ancona NP sample from day 21, displayed in Fig. 3 of the main text. The subset consists of 18 genes involved in uric acid metabolism from a total of 32,180 ORFs identified in the metagenome. Gene sequences with similarity to Halomonas spp. are printed in bold. (XLSX 10 kb)
Supplementary Table 3d
Annotation of the metagenomic data from the Bizerte NP sample from day 21, displayed in Fig. 3 of the main text. The subset consists of six genes involved in uric acid metabolism from a total of 28,698 ORFs identified in the metagenome. Gene sequences with similarity to Halomonas spp. are printed in bold. (XLSX 9 kb)
Supplementary Table 3e
Annotation of the metagenomic data from the El Max UA sample from day 21, displayed in Fig. 3 of the main text. The subset consists of 29 genes involved in uric acid metabolism from a total of 61,277 ORFs identified in the metagenome. Gene sequences with similarity to Halomonas spp. are printed in bold. (XLSX 10 kb)
Supplementary Table 4
Closest relatives of 16S rRNA gene sequences of uric acid-degrading isolates. The first letter of the isolate code designates the sediment sampling location (A—Ancona; Q—Aqaba; X—El Max; B—Bizerte), and the second letter indicates the nitrogen source (N—Ammonium; U—Uric Acid). The first number in the isolate code represents the sampling date, and the last number indicates the individual isolate. (XLSX 10 kb)
Supplementary Fig. 1
RISA fingerprinting profiles used for the multivariate analysis presented in Fig. 2 of the main text. Full DNA profiles of two out of three microcosm replicates were used to avoid mismatching in the band matching required for statistical analysis. All RISA fingerprints were run using the O’gene Ruler Plus (Thermo Scientific, Lutterworth, UK) as indicated by the letter M under the marker lanes. Gel images show DNA fragments of 300 to 5000 bp in length. Numbers under individual lanes represent sampling days. Fingerprints from microcosm replicates are displayed as blocks of profiles with increasing sampling time (0 to 28 days). (GIF 304 kb)
Rights and permissions
About this article
Cite this article
Gertler, C., Bargiela, R., Mapelli, F. et al. Conversion of Uric Acid into Ammonium in Oil-Degrading Marine Microbial Communities: a Possible Role of Halomonads. Microb Ecol 70, 724–740 (2015). https://doi.org/10.1007/s00248-015-0606-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-015-0606-7