Abstract
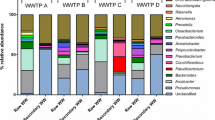
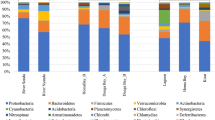
Microbial communities from two field-scale swine wastewater treatment plants (WWTPs) were assessed by pyrosequencing analyses of bacterial and archaeal 16S ribosomal DNA (rDNA) fragments. Effluent samples from secondary (anaerobic covered lagoons and upflow anaerobic sludge blanket [UASB]) and tertiary treatment systems (open-pond natural attenuation lagoon and air-sparged nitrification-denitrification tank followed by alkaline phosphorus precipitation process) were analyzed. A total of 56,807 and 48,859 high-quality reads were obtained from bacterial and archaeal libraries, respectively. Dominant bacterial communities were associated with the phylum Firmicutes, Bacteroidetes, Proteobacteria, or Actinobacteria. Bacteria and archaea diversity were highest in UASB effluent sample. Escherichia, Lactobacillus, Bacteroides, and/or Prevotella were used as indicators of putative pathogen reduction throughout the WWTPs. Satisfactory pathogen reduction was observed after the open-pond natural attenuation lagoon but not after the air-sparged nitrification/denitrification followed by alkaline phosphorus precipitation treatment processes. Among the archaeal communities, 80 % of the reads was related to hydrogeno-trophic methanogens Methanospirillum. Enrichment of hydrogenotrophic methanogens detected in effluent samples from the anaerobic covered lagoons and UASB suggested that CO2 reduction with H2 was the dominant methanogenic pathway in these systems. Overall, the results served to improve our current understanding of major microbial communities’ changes downgradient from the pen and throughout swine WWTP as a result of different treatment processes.




Similar content being viewed by others
References
ABIPECS (2011) Annual report. Associacao Brasileira da Industria Produtora e Exportadora de Carne Suina. http://www.abipecs.org.br/uploads/relatorios/relatorios-associados-ingles/Abipecs_annual_report_2011.pdf. Accessed 07 July 2014
Cronk JK (1996) Constructed wetlands to treat wastewater from dairy and swine operations: a review. Agric Ecosyst Environ 58:97–114. doi:10.1016/0167-8809(96)01024-9
Viancelli A, Kunz A, Steinmetz RLR, Kich JD, Souza CK, Canal CW, Coldebella A, Esteves PA, Barardi CRM (2013) Performance of two swine manure treatment systems on chemical composition and on the reduction of pathogens. Chemosphere 90:1539–1544. doi:10.1016/j.chemosphere.2012.08.055
Jeong JY, Park HD, Lee KH, Weon HY, Ka JO (2011) Microbial community analysis and identification of alternative host-specific fecal indicators in fecal and river water samples using pyrosequencing. J Microbiol (Seoul, Korea) 49:585–594. doi:10.1007/s12275-011-0530-6
Lamendella R, Santo Domingo JW, Yannarell AC, Ghosh S, Di Giovanni G, Mackie RI, Oerther DB (2009) Evaluation of swine-specific PCR assays used for fecal source tracking and analysis of molecular diversity of swine-specific “bacteroidales” populations. Appl Environ Microbiol 75:5787–5796. doi:10.1128/aem. 00448-09
Goh SH, Mabbett AN, Welch JP, Hall SJ, McEwan AG (2009) Molecular ecology of a facultative swine waste lagoon. Lett Appl Microbiol 48:486–492. doi:10.1111/j.1472-765X.2009.02560.x
Ducey TF, Hunt PG (2013) Microbial community analysis of swine wastewater anaerobic lagoons by next-generation DNA sequencing. Anaerobe 21:50–57. doi:10.1016/j.anaerobe.2013.03.005
Peu P, Brugere H, Pourcher AM, Kerouredan M, Godon JJ, Delgenes JP, Dabert P (2006) Dynamics of a pig slurry microbial community during anaerobic storage and management. Appl Environ Microbiol 72:3578–3585. doi:10.1128/aem. 72.5.3578-3585.2006
Snell-Castro R, Godon JJ, Delgenes JP, Dabert P (2005) Characterisation of the microbial diversity in a pig manure storage pit using small subunit rDNA sequence analysis. FEMS Microbiol Ecol 52:229–242. doi:10.1016/j.femsec.2004.11.016
Kim W, Lee S, Shin SG, Lee C, Hwang K, Hwang S (2010) Methanogenic community shift in anaerobic batch digesters treating swine wastewater. Water Res 44:4900–4907. doi:10.1016/j.watres.2010.07.029
Muhlbauer RV, Moody LB, Burns RT, Harmon J, Stalder K (2010) Water consumption and conservation techniques currently available for swine production. National Pork Board, pp. 32
Federation WE, Association APH (2005) Standard methods for the examination of water and wastewater. American Public Health Association (APHA), Washington
Nossa CW, Oberdorf WE, Yang L, Aas JA, Paster BJ, Desantis TZ, Brodie EL, Malamud D, Poles MA, Pei Z (2010) Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J Gastroenterol: WJG 16:4135–4144
Gantner S, Andersson AF, Alonso-Saez L, Bertilsson S (2011) Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J Microbiol Methods 84:12–18. doi:10.1016/j.mimet.2010.10.001
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi:10.1128/aem. 01541-09
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6:e27310. doi:10.1371/journal.pone.0027310
Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6:639–641. doi:10.1038/nmeth.1361
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi:10.1038/nmeth.f.303
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinforma (Oxford, England) 27:2194–2200. doi:10.1093/bioinformatics/btr381
Chao A, Shen T-J (2003) Nonparametric estimation of Shannon’s index of diversity when there are unseen species in sample. Environ Ecol Stat 10:429–443. doi:10.1023/A:1026096204727
Chao A, Shen T-J (2010) Program SPADE (Species Predition and Diversity Estimation)
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264. doi:10.1093/biomet/40.3-4.237
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press
Yue JC, Clayton MK (2005) A similarity measure based on species proportions. Commun Stat-Theory Methods 34:2123–2131
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Parks DH, Beiko RG (2010) Identifying biologically relevant differences between metagenomic communities. Bioinforma (Oxford, England) 26:715–721. doi:10.1093/bioinformatics/btq041
Chong S, Sen TK, Kayaalp A, Ang HM (2012) The performance enhancements of upflow anaerobic sludge blanket (UASB) reactors for domestic sludge treatment—a state-of-the-art review. Water Res 46:3434–3470. doi:10.1016/j.watres.2012.03.066
Cotta MA, Whitehead TR, Collins MD, Lawson PA (2004) Atopostipes suicloacale gen. nov., sp. nov., isolated from an underground swine manure storage pit. Anaerobe 10:191–195. doi:10.1016/j.anaerobe.2004.04.001
Pagnier I, Croce O, Robert C, Raoult D, La Scola B (2013) Non-contiguous finished genome sequence and description of Anaerococcus pacaensis sp. nov., a new species of anaerobic bacterium. Stand Genomic Sci 8:548–560. doi:10.4056/sigs.4177252
Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S (2001) Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol 51:1521–1528
Yang J, Martinez I, Walter J, Keshavarzian A, Rose DJ (2013) In vitro characterization of the impact of selected dietary fibers on fecal microbiota composition and short chain fatty acid production. Anaerobe 23:74–81. doi:10.1016/j.anaerobe.2013.06.012
Palatsi J, Illa J, Prenafeta-Boldu FX, Laureni M, Fernandez B, Angelidaki I, Flotats X (2010) Long-chain fatty acids inhibition and adaptation process in anaerobic thermophilic digestion: batch tests, microbial community structure and mathematical modelling. Bioresour Technol 101:2243–2251. doi:10.1016/j.biortech.2009.11.069
Lin L, Wan C, Liu X, Lee DJ, Lei Z, Zhang Y, Tay JH (2013) Effect of initial pH on mesophilic hydrolysis and acidification of swine manure. Bioresour Technol 136:302–308. doi:10.1016/j.biortech.2013.02.106
Syutsubo K, Nagaya Y, Sakai S, Miya A (2005) Behavior of cellulose-degrading bacteria in thermophilic anaerobic digestion process. Water Sci Technol J Int Assoc Water Pollut Res 52:79–84
Madigan MT, Martinko JM, Parker J (2000) Brock biology of microorganisms. Prentice Hall, New Jersey
Walter J (2008) Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol 74:4985–4996. doi:10.1128/aem. 00753-08
Ziemer CJ (2013) Broad diversity and newly cultured bacterial isolates from enrichment of pig feces on complex polysaccharides. Microb Ecol 66:448–461. doi:10.1007/s00248-013-0185-4
Hanreich A, Schimpf U, Zakrzewski M, Schluter A, Benndorf D, Heyer R, Rapp E, Puhler A, Reichl U, Klocke M (2013) Metagenome and metaproteome analyses of microbial communities in mesophilic biogas-producing anaerobic batch fermentations indicate concerted plant carbohydrate degradation. Syst Appl Microbiol 36:330–338. doi:10.1016/j.syapm.2013.03.006
Rettedal E, Vilain S, Lindblom S, Lehnert K, Scofield C, George S, Clay S, Kaushik RS, Rosa AJ, Francis D, Brozel VS (2009) Alteration of the ileal microbiota of weanling piglets by the growth-promoting antibiotic chlortetracycline. Appl Environ Microbiol 75:5489–5495. doi:10.1128/aem. 02220-08
Egert M, Stingl U, Bruun LD, Pommerenke B, Brune A, Friedrich MW (2005) Structure and topology of microbial communities in the major gut compartments of Melolontha larvae (Coleoptera: Scarabaeidae). Appl Environ Microbiol 71:4556–4566. doi:10.1128/aem. 71.8.4556-4566.2005
McGarvey JA, Miller WG, Sanchez S, Stanker L (2004) Identification of bacterial populations in dairy wastewaters by use of 16S rRNA gene sequences and other genetic markers. Appl Environ Microbiol 70:4267–4275. doi:10.1128/aem. 70.7.4267-4275.2004
Delbes C, Ali-Mandjee L, Montel MC (2007) Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl Environ Microbiol 73:1882–1891. doi:10.1128/aem. 01716-06
Bosshard PP, Zbinden R, Altwegg M (2002) Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, Gram-positive bacterium. Int J Syst Evol Microbiol 52:1263–1266
Sohaskey CD, Modesti L (2009) Differences in nitrate reduction between Mycobacterium tuberculosis and Mycobacterium bovis are due to differential expression of both narGHJI and narK2. FEMS Microbiol Lett 290:129–134. doi:10.1111/j.1574-6968.2008.01424.x
Khan A, Sarkar D (2012) Nitrate reduction pathways in mycobacteria and their implications during latency. Microbiol (Reading, England) 158:301–307
Kay D, Crowther J, Stapleton CM, Wyer MD, Fewtrell L, Edwards A, Francis CA, McDonald AT, Watkins J, Wilkinson J (2008) Faecal indicator organism concentrations in sewage and treated effluents. Water Res 42:442–454. doi:10.1016/j.watres.2007.07.036
Topp E, Scott A, Lapen DR, Lyautey E, Duriez P (2009) Livestock waste treatment systems for reducing environmental exposure to hazardous enteric pathogens: some considerations. Bioresour Technol 100:5395–5398. doi:10.1016/j.biortech.2008.11.001
Brooks JP, Adeli A, McLaughlin MR (2014) Microbial ecology, bacterial pathogens, and antibiotic resistant genes in swine manure wastewater as influenced by three swine management systems. Water Res 57:96–103. doi:10.1016/j.watres.2014.03.017
Bernhard AE, Field KG (2000) Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl Environ Microbiol 66:1587–1594. doi:10.1128/aem. 66.4.1587-1594.2000
Bower PA, Scopel CO, Jensen ET, Depas MM, McLellan SL (2005) Detection of genetic markers of fecal indicator bacteria in Lake Michigan and determination of their relationship to Escherichia coli densities using standard microbiological methods. Appl Environ Microbiol 71:8305–8313
Mauffret A, Caprais MP, Gourmelon M (2012) Relevance of Bacteroidales and F-specific RNA bacteriophages for efficient fecal contamination tracking at the level of a catchment in France. Appl Environ Microbiol 78:5143–5152. doi:10.1128/AEM. 00315-12
Okabe S, Okayama N, Savichtcheva O, Ito T (2007) Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl Microbiol Biotechnol 74:890–901. doi:10.1007/s00253-006-0714-x
Kant R, Paulin L, Alatalo E, de Vos WM, Palva A (2011) Genome sequence of Lactobacillus amylovorus GRL1118, isolated from pig ileum. J Bacteriol 193:3147–3148. doi:10.1128/JB.00423-11
Marti R, Dabert P, Ziebal C, Pourcher A-M (2010) Evaluation of Lactobacillus sobrius/L. amylovorus as a new microbial marker of pig manure. Appl Environ Microbiol 76:1456–1461. doi:10.1128/aem. 01895-09
Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R (1997) Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol 63:2802–2813
Fu B, Jiang Q, Liu H, Liu H (2014) Occurrence and reactivation of viable but non-culturable E. coli in sewage sludge after mesophilic and thermophilic anaerobic digestion. Biotechnol Lett 36:273–279. doi:10.1007/s10529-013-1361-9
Vanotti MB, Millner PD, Hunt PG, Ellison AQ (2005) Removal of pathogen and indicator microorganisms from liquid swine manure in multi-step biological and chemical treatment. Bioresour Technol 96:209–214. doi:10.1016/j.biortech.2004.05.010
Wong JWC, Selvam A (2009) Reduction of indicator and pathogenic microorganisms in pig manure through fly ash and lime addition during alkaline stabilization. J Hazard Mater 169:882–889. doi:10.1016/j.jhazmat.2009.04.033
Worm P, Stams AJ, Cheng X, Plugge CM (2011) Growth- and substrate-dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei. Microbiol (Reading, England) 157:280–289. doi:10.1099/mic.0.043927-0
Chauhan A, Ogram A, Reddy KR (2004) Syntrophic-methanogenic associations along a nutrient gradient in the Florida Everglades. Appl Environ Microbiol 70:3475–3484. doi:10.1128/aem. 70.6.3475-3484.2004
Ferry JG, Smith PH, Wolfe R (1974) Methanospirillum, a new genus of methanogenic bacteria, and characterization of Methanospirillum hungatii sp. nov. Int J Syst Bacteriol 24:465–469
Ollivier BM, Mah RA, Garcia J, Boone DR (1986) Isolation and characterization of Methanogenium bourgense sp. nov. Int J Syst Bacteriol 36:297–301
Talbot G, Roy C, Topp E, Kalmokoff M, Brooks S, Beaulieu C, Palin M, Masse D (2010) Spatial distribution of some microbial trophic groups in a plug-flow-type anaerobic bioreactor treating swine manure
Miller TL, Lin C (2002) Description of Methanobrevibacter gottschalkii sp. nov., Methanobrevibacter thaueri sp. nov., Methanobrevibacter woesei sp. nov. and Methanobrevibacter wolinii sp. nov. Int J Syst Evol Microbiol 52:819–822
Ike M, Inoue D, Miyano T, Liu TT, Sei K, Soda S, Kadoshin S (2010) Microbial population dynamics during startup of a full-scale anaerobic digester treating industrial food waste in Kyoto eco-energy project. Bioresour Technol 101:3952–3957
Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D (2002) The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res 12:532–542
Liu Y, Whitman WB (2008) Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci 1125:171–189
Zhang C, Yuan Q, Lu Y (2014) Inhibitory effects of ammonia on methanogen mcrA transcripts in anaerobic digester sludge. FEMS Microbiol Ecol 87:368–377
Fotidis IA, Karakashev D, Angelidaki I (2013) Bioaugmentation with an acetate-oxidising consortium as a tool to tackle ammonia inhibition of anaerobic digestion. Bioresour Technol 146:57–62. doi:10.1016/j.biortech.2013.07.041
Briones A, Raskin L (2003) Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr Opin Biotechnol 14:270–276
Kovacik WP, Scholten JC, Culley D, Hickey R, Zhang W, Brockman FJ (2010) Microbial dynamics in upflow anaerobic sludge blanket (UASB) bioreactor granules in response to short-term changes in substrate feed. Microbiol (Reading, England) 156:2418–2427
Fernandes GW, Kunz A, Steinmetz RL, Szogi A, Vanotti M, Flores EM, Dressler VL (2012) Chemical phosphorus removal: a clean strategy for piggery wastewater management in Brazil. Environ Technol 33:1677–1683. doi:10.1080/09593330.2011.642896
Kunz A, Miele M, Steinmetz RL (2009) Advanced swine manure treatment and utilization in Brazil. Bioresour Technol 100:5485–5489. doi:10.1016/j.biortech.2008.10.039
Acknowledgments
This research was supported by the Brazilian Agricultural Research Corporation (EMBRAPA) grant no. 01.11.07.002.05.00.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Da Silva, M.L.B., Cantão, M.E., Mezzari, M.P. et al. Assessment of Bacterial and Archaeal Community Structure in Swine Wastewater Treatment Processes. Microb Ecol 70, 77–87 (2015). https://doi.org/10.1007/s00248-014-0537-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0537-8