Abstract
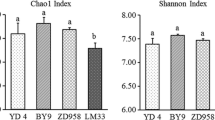
Rhizosphere microorganisms in soils are important for plant growth. However, the importance of rhizosphere microorganisms is still underestimated since many microorganisms associated with plant roots cannot be cultured and since the microbial diversity in the rhizosphere can be influenced by several factors, such as the cropping history, biogeography, and agricultural practice. Here, we characterized the rhizosphere bacterial diversity of cucumber plants grown in soils covering a wide range of cucumber cropping histories and environmental conditions by using pyrosequencing of bacterial 16S rRNA genes. We also tested the effects of compost addition and/or bacterial inoculation on the bacterial diversity in the rhizosphere. We identified an average of approximately 8,883 reads per sample, corresponding to around 4,993 molecular operational taxonomic units per sample. The Proteobacteria was the most abundant phylum in almost all soils. The abundances of the phyla Bacteroidetes, Actinobacteria, Firmicutes, Acidobacteria, and Verrucomicrobia varied among the samples, and together with Proteobacteria, these phyla were the six most abundant phyla in almost all analyzed samples. Analyzing all the sample libraries together, the predominant genera found were Flavobacterium, Ohtaekwangia, Opitutus, Gp6, Steroidobacter, and Acidovorax. Overall, compost and microbial amendments increased shoot biomass when compared to untreated soils. However, compost addition decreased the bacterial α-diversity in most soils (but for three soils compost increased diversity), and no statistical effect of microbial amendment on the bacterial α-diversity was found. Moreover, soil amendments did not significantly influence the bacterial β-diversity. Soil organic content appeared more important than compost and microbial amendments in shaping the structure of bacterial communities in the rhizosphere of cucumber.







Similar content being viewed by others
References
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266
Jackson LE, Bowles TM, Hodson AK, Lazcano C (2012) Soil microbial-root and microbial-rhizosphere processes to increase nitrogen availability and retention in agroecosystems. Curr Opin Environ Sustain 4:517–522
Schenk PM, Carvalhais LC, Kazan K (2012) Unraveling plant–microbe interactions: can multi-species transcriptomics help? Trends Biotechnol 30:177–184
Berendsen RL, Pieterse CM, Bakker PA (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Newton AC, Fitt BD, Atkins SD, Walters DR, Daniell TJ (2010) Pathogenesis, parasitism and mutualism in the trophic space of microbe–plant interactions. Trends Microbiol 18:365–373
Teixeira LC, Peixoto RS, Cury JC, Sul WJ, Pellizari VH, Tiedje J, Rosado AS (2010) Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J 4:989–1001
Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JH et al (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100
Buée M, De Boer W, Martin F, Van OL, Jurkevitch E (2009) The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189–212
Mazurier S, Corberand T, Lemanceau P, Raaijmakers JM (2009) Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J 3:977–991
Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F et al (2008) Stress tolerance in plants via habitat-adapted symbiosis. ISME J 2:404–416
Morris RM, Nunn BL, Frazar C, Goodlett DR, Ting YS, Rocap G (2010) Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J 4:673–685
Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J et al (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230
Bressan M, Roncato MA, Bellvert F, Comte G, Haichar ZF, Achouak W, Berge O (2009) Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J 3:1243–1257
DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE, Firestone MK (2008) Selective progressive response of soil microbial community to wild oat roots. ISME J 3:168–178
Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F et al (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S et al (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90
Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, Swarbreck D et al (2013) Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J 7:2248–2258
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL et al (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110:6548–6553
Bennett AJ, Bending GD, Chandler DC, Hilton S, Mills P (2012) Meeting the demand for crop production: the challenge of yield decline in crops grown in short rotations. Biol Rev 87:52–71
Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG et al (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351
Reeve JR, Schadt CW, Carpenter-Boggs L, Kang S, Zhou J, Reganold JP (2010) Effects of soil type and farm management on soil ecological functional genes and microbial activities. ISME J 4:1099–1107
Jesus ED, Marsh TL, Tiedje JM, Moreira FMD (2009) Changes in lands use alter the structure of bacterial communities in Western Amazon soils. ISME J 3:1004–1011
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 57:5111–5120
Huang S, Li R, Zhang Z, Li L, Gu X, Fan W et al (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41:1275–1281
Yu JQ, Ye SF, Zhang MF, Hu WH (2003) Effects of root exudates and aqueous root extracts of cucumber (Cucumis sativus) and allelochemicals, on photosynthesis and antioxidant enzymes in cucumber. Biochem Syst Ecol 31:129–139
Ye SF, Yu JQ, Peng YH, Zheng JH, Zou LY (2004) Incidence of Fusarium wilt in Cucumis sativus L. is promoted by cinnamic acid, an autotoxin in root exudates. Plant Soil 263:143–150
Zhou X, Wu F (2012) Effects of amendments of ferulic acid on soil microbial communities in the rhizosphere of cucumber (Cucumis sativus L.). Eur J Soil Biol 50:191–197
Ibekwe AM, Poss JA, Grattan SR, Grieve CM, Suarez D (2010) Bacterial diversity in cucumber (Cucumis sativus) rhizosphere in response to salinity, soil pH, and boron. Soil Biol Biochem 42:567–575
Zhou X, Wu F (2013) Artificially applied vanillic acid changed soil microbial communities in the rhizosphere of cucumber (Cucumis sativus L.). Can J Soil Sci 93:13–21
Chung S, Kong H, Buyer JS, Lakshman DK, Lydon J, Kim S-D, Roberts DP (2008) Isolation and partial characterization of Bacillus subtilis ME488 for suppression of soilborne pathogens of cucumber and pepper. Appl Microbiol Biotechnol 280:115–123
Ofek M, Hadar Y, Minz D (2011) Colonization of cucumber seeds by bacteria during germination. Environ Microbiol 13:2794–2807
Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH (2008) Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol 17:1636–1647
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123
Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R (2011) Experimental and analytical tools for studying the human microbiome. Nat Rev Genet 13:47–58
Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P (2010) Experimental factors affecting PCR based estimates of microbial species richness and evenness. ISME J 4:642–647
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Tian Y, Zhang X, Liu J, Gao L (2011) Effects of summer cover crop and residue management on cucumber growth in intensive Chinese production systems: soil nutrients, microbial properties and nematodes. Plant Soil 339:299–315
Shanon CE, Weaver W (1949) The mathematical theory of communications. University of Illinois Press, Urbana
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, et al (2013) Package ‘Vegan’, Community Ecology Package. Version 2.0-8. http://cran.r-project.org. Accessed July 10, 2013
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728
Mahaffee WF, Kloepper JW (1997) Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field-grown cucumber (Cucumis sativus L.). Microb Ecol 34:210–223
Zhou X, Wu F (2012) p-Coumaric ccid influenced cucumber rhizosphere soil microbial communities and the growth of Fusarium oxysporum f. sp. cucumerinum owen. PLoS One 7:e48288
Xu Y, Wang G, Jin J, Liu J, Zhang Q, Liu X (2009) Bacterial communities in soybean rhizosphere in response to soil type, soybean genotype, and their growth stage. Soil Biol Biochem 41:919–925
Uroz S, Buee M, Murat C, Frey-Klett P, Martin F (2010) Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep 2:281–288
Gottel NR, Castro HF, Kerley M, Yang Z, Pelletier DA, Podar M et al (2011) Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl Environ Microbiol 77:5934–5944
Spain AM, Krumholz LR, Elshahed MS (2009) Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J 3:992–1000
Naumoff DG, Dedysh SN (2012) Lateral gene transfer between the Bacteroidetes and Acidobacteria: the case of α-L-Rhamnosidases. FEBS Lett 586:3843–3851
Leggett MJ, McDonnell G, Denyer SP, Setlow P, Maillard JY (2012) Bacterial spore structures and their protective role in biocide resistance. J Appl Microbiol 113:485–498
Qin S, Xing K, Jiang JH, Xu LH, Li WJ (2011) Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl Microbiol Biotechnol 89:457–473
Kim JS, Dungan RS, Kwon SW, Weon HY (2006) The community composition of root-associated bacteria of the tomato plant. World J Microbiol Biotechnol 22:1267–1273
Manter DK, Delgado JA, Holm DG, Stong RA (2010) Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microb Ecol 60:157–166
Germida JJ, Siciliano SD, de Freitas JR, Seib AM (1998) Diversity of root-associated bacteria associated with held-grown canola (Brassica napus L.) and wheat (Triticum aestivum L.). FEMS Microbiol Ecol 26:43–50
Bernardet JF, Bowman JP (2006) The genus Flavobacterium. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes: a handbook on the biology of bacteria, vol 7, 3rd edn. Springer, New York, pp 481–531
Tejeda-Agredano MC, Gallego S, Vila J, Grifoll M, Ortega-Calvo JJ, Cantos M (2013) Influence of the sunflower rhizosphere on the biodegradation of PAHs in soil. Soil Biol Biochem 57:830–840
Zhang YZ, Wang ET, Li M, Li QQ, Zhang YM, Zhao SJ et al (2011) Effects of rhizobial inoculation, cropping systems and growth stages on endophytic bacterial community of soybean roots. Plant Soil 347:147–161
Gaggìa F, Baffoni L, Di Gioia D, Accorsi M, Bosi S, Marotti I et al (2013) Inoculation with microorganisms of Lolium perenne L.: evaluation of plant growth parameters and endophytic colonization of roots. New Biotechnol 30:695–704
Acknowledgments
We are grateful to the Chinese Universities Scientific Fund (2014JD047 and 2014XJ035) and Beijing Innovation Team for Fruity Vegetables in Chinese Modern Agricultural Technology System for their financial supports.
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 2126 kb)
Rights and permissions
About this article
Cite this article
Tian, Y., Gao, L. Bacterial Diversity in the Rhizosphere of Cucumbers Grown in Soils Covering a Wide Range of Cucumber Cropping Histories and Environmental Conditions. Microb Ecol 68, 794–806 (2014). https://doi.org/10.1007/s00248-014-0461-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0461-y