Abstract
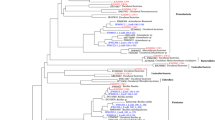
Bacteria were identified from a large, seasonally flooded river (Paraná River, Brazil) and two floodplain habitats that were part of the same river system yet very different in nature: clearwater Garças Lagoon and the highly humic waters of Patos Lagoon. Bacterioplankton were collected during mid-summer (Jan. 2002) from water samples (2 l) filtered first through a 1.2-μm filter then a 0.2-μm membrane filter representing the particle-attached and free-living sub-communities, respectively. DNA was extracted from filters and purified and a 16S rRNA clone library established for each habitat. Over 300 clones were sequenced and checked for similarity to existing 16S sequences in GenBank using the BLAST algorithm with default parameters. Further classification of clones was done using a species “backbone” attachment followed by parsimony analysis. The majority (85%) of sequences, referred to here as operational taxonomic units (OTUs), were most similar to uncultured bacterium 16S sequences. OTUs from each Proteobacteria sub-phylum (α, β, γ, δ, ɛ) were present in the Upper Paraná River system, as well as members of the Bacteroidetes. The microbial assemblage from Patos Lagoon was least like other samples in that it had no Firmicutes present and was dominated by Actinobacteria. Verrucomicrobia OTUs were only found in the free-living assemblage. This study documents the presence of globally distributed phyla in Upper Paraná River and taxa unique to habitat and particle attachment.




Similar content being viewed by others
References
Alfreider A, Pernthaler J, Amann R, Sattler B, Glockner F, Wille A, Psenner R (1996) Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol 62:2138–2144
Agostinho AA, Thomaz SM, Minte-Vera CV, Winemiller KO (2000) Biodiversity in the High River Paraná floodplain. In: Gopal B, Junk WJ, Davis JA (eds) Biodiversity in wetlands: assessment, function and conservation. Backhuys, Leiden, pp 89–118
Agostinho AA, Zalewski M (1996) A planicie alagavel do alto rio Paraná: Importancia e preservacao. EDUEM, Maringa
Bahr M, Hobbie JE, Sogin ML (1996) Bacterial diversity in an arctic lake—a freshwater SAR1 cluster. Aquat Microb Ecol 11:271–277
Barreto SRG, Nozaki J, Barreto WJ (2003) Origin of dissolved organic carbon studied by UV–vis spectroscopy. Acta Hydrochimmica et Hydrobiologica 31:513–518
Besemer KM, Moeseneder M, Arrieta JM, Herndl GJ, Peduzzi P (2005) Complexity of bacterial communities in a river-floodplain system (Danube, Austria). Appl Environ Microbiol 71:609–620
Borneman J, Skroch PW, O’Sullivan KM, Palus JA, Rumjanek NG, Jansen JL, Nienhuis J, Triplett EW (1996) Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol 62:1935–1943
Bosshard PP, Santini Y, Grütter D, Stettler R, Bachofen R (2000) Bacterial diversity and community composition in the chemocline of the meromictic alpine Lake Cadagno as revealed by 16S rDNA analysis. FEMS Microbial Ecol 31:173–182
Bukert U, Warnecke F, Babenzien D, Zwirnmann E, Pernthaler J (2003) Members of a readily enriched β-proteobacterial clade are common in surface waters of a humic lake. Appl Environ Microbio. 69:6550–6559
Carvalho P, Thomaz SM, Bini LM (2003) Effects of water level, abiotic and biotic factors on bacterioplankton abundance in lagoons of a tropical flooplain (Paraná River, Brazil). Hydrobiologia 510:67–74
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Crump BC, Baross JA (1996) Particle-attached bacteria and heterotrophic plankton in the Columbia River estuary. Mar Ecol Prog Ser 138:265–273
Crump BC, Baross JA (2000) Characterization of the bacterially-active particle fraction in the Columbia River estuary. Mar Ecol Prog Ser 206:12–22
Crump BC, Imenstad CA, Baross JA (1998) Particle-attached bacteria dominant the Columbia River estuary. Aquat Microb Ecol 14:7–18
Crump BC, Armbrust EV, Baross JA (1999) Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol 65:3192–3204
Crump BC, Kling GW, Bahr M, Hobbie JE (2003) Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl Environ Microbiol 69:2253–2268
Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (Eds) (2006) The prokaryotes, 3rd ed. Springer, NY
Eiler A, Bertilsson S (2004) Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ Microbiol 6:1228–1243
Felsenstein J (1978) Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool 27:401–410
Gine MF, Bergamin H, Zaggato EAG, Reis BF (1980) Simultaneous determination of nitrate and nitrite by flow-injection analysis. Analytica Chimica Acta 114:191–197
Glöckner FO, Fuchs BM, Amann R (1999) Bacterioplankton compositions of lakes and oceans: A first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol 65:3721–3726
Glöckner FO, Zaichikov E, Belkova N, Denissova L, Pernthaler J, Pernthaler A, Amann R (2000) Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Appl Environ Microbiol 66:5053–5065
Golterman HL, Clymo RS, Ohmstad MAM (1978) Methods for physical and chemical analysis of fresh waters. Blackwell Scientific, Oxford, p 214
Haukka K, Heikkinen E, Kairesalo T, Karjaiainen H, Sivoneh K (2005) Effect of humic material on the bacterioplankton community composition in boreal lakes and mesocosms. Environ Microbiol 7:620–630
Hiorns WD, Methe BA, Nierzwicki-Bauer SA, Zehr JP (1997) Bacterial diversity in Adirondack mountain lakes as reveled by 16S rRNA gene sequences. Appl Environ Microiol 63:2957–2960
Junk WJ, Bayley PB, Sparks RE (1989) The floodpluse concept in river-floodplain systems. In: Dodge DP (ed) Proceedings of the International Large River Symposium, pp 110–127. Can. Spec. Publ. Fish. Aquat. Sci. 106
Knapp JS (1988) Historical perspectives and identification of Neisseria and related species. Clin Microbiol Rev 1:415–431
Lane DJ (1991) 16S/23S rRNA Sequencing. In: Stackebrandt E, Goofellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lindström ES (2000) Bacterioplankton community composition in a boreal forest lake. FEMS Microbiol Ecol 27:163–174
Lyautey E, Lacoste B, Ten-Hage L, Rols J-L, Garabetian F (2005) Analysis of bacterial diversity in river biofilms using 16S rDNA PCR-DGGE: Methodological settings and fingerprints interpretation. Water Res 39:380–388
Mackereth FYH, Heron J, Talling JJ (1978) Water analysis: some revised methods for limnologists. Freshwater Biological Association. Scientific Publication No. 36, Cumbria, p 120
Maddison DR, Maddison WP (2001) MacClade analysis of phylogeny and character evolution, Ver. 4. Wayne P. Sinauer, Sunderland, MA
Maidak BL, Cole JR, Liburn TG, Parker CT, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM (2001) The RDP-II (Ribosomal Database Project). Nucleic Acid Res 29:173–174
Methé BA, Hiorns WD, Zehr JP (1998) Contrasts between marine and freshwater bacterial community composition—analyses of communities in Lake George and six other Adirondack lakes. Limnol Oceanogr 43:368–374
Methé BA, Zehr JP (1999) Diversity of bacterial communities in Adirondack lakes: do species assemblages reflect lake water chemistry? Hydrobiologia 401:77–96
Muyzer G, De Waal ED, Uitterlindiden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nakatsu CH, Torsvik V, Øveås L (2000) Soil community analysis using DGGE of 16S rDNA polymerase chain reaction products. Soil Sci Soc Am J 64:1382–1388
Oliveira JL, Boroski M, Azevedo JCR, Nozaki J (2006) Spectroscopic investigation of humic substances in a tropical lake during a complete hydrological cycle. Acta Hydrochimmica et Hydrobiologica 34:608–617
Rodrigues LC, Train S, do C, Roberto M, Pagioro TA (2002) Seasonal fluctuation of some limnological variables on a floodplain lake (Patos lagoon) of the Upper Paraná River, Mato Grosso do Sul State, Brazil. Brazilian Arch of Biol Tech 45:499–513
Simestad CA, Morgan CA, Cordell JR, Baross JA (1994) Flux, passive retention, and active residence of zooplankton in Columbia River estuarine turbidity maxima. In: Dyer KR, Orth BJ (eds) Changes in fluxes in estuaries: implications from science to management (ECSA22/ERF symposium, Plymouth, September 1992). Olsen and Olsen, Fredensborg, Denmark, pp 47–484
Sarkar N, Thornton JW, Planet PJ, Figurski DH, Schierwater D, DeSalle R (2002) An automated phylogenetic key for classifying homeoboxes. Mol Phylogenet Evol 24:388–399
Souza FEE, Stevaux JC (1997) Geolgia e geomorfologia do complex rio Baia, Corutuba, Ivinhema. In: Vazzoler AEAM, Agostinho AA, Hahn NS (eds) A Planicie de inundacao do alto rio Paraná: Aspectos Fisico Biologicos e socioeconomicos. EDUEM, Maringa, pp 3–43
Swofford DL (2002) Phylogenetic analysis using parsimony* version 4. Wayne P. Sinauer, Sunderland, MA
Thomaz SM, Pagioro TA, Bini LM, do Roberto CM, de Araújo Rocha RR (2004) Chapter 4: Limnological characterization of the aquatic environments and the influence of hydrometric levels. In: Thomaz SM, Agostinho AA, Hahn NS (eds) The upper Paraná River and its flooplain: physical aspects, ecology and conservation. Backhuys, Leiden, The Netherlands, pp 75–102
Thomaz S, Bini L, Bozelli R (2007) Floods increase similarity among aquatic habitats in river-flooplain systems. Hydrobiologia 579:1–13
Van der Gucht K, Sabbe K, De Meester L, Vloemans N, Zwart G, Gillis M, Vyverman W (2001) Contrasting bacterioplankton community composition and seasonal dynamics in two neighbouring hypertrophic freshwater lakes. Environ Microbiol 3:680–690
Ward N, Rainey FA, Stackebrandt E, Schlesner H (1995) Unraveling the extent of diversity within the order Planctomycetales. Appl Environ Microbiol 61:2270–2275
Warnecke R, Amann R, Pernthaler J (2004) Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ Microbiol 6:242–253
Weiss P, Schweitzer B, Amann R, Simon M (1996) Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow). Appl Envion Microbiol 62:1998–2005
Winter C, Hein T, Kavka G, Mach RL, Farnleitner AH (2007) Longitudinal changes in the bacterial community composition of the Danube River: a whole-river approach. Appl Environ Microbiol 73:421–431
Zavarzin GA, Stackebrandt E, Murray RG (1991) A correlation of phylogenetic diversity in Proteobacteria with the influences of ecological forces. Can J Microbiol 37:1–6
Zwart G, Crump BC, Kamst-van Agterveld M, Hagen F, Han S-K (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microbial Ecol 28:141–155
Zwart G, Hiorns WD, Methe BA, van Agterveld MP, Huismans R, Nold SC, Zehr JP, Laanbroek HJ (1998) Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst Appl Microbiol 21:546–556
Zwart G, Huismans R, van Agterveld MP, Van de Peer Y, De Rijk P, Eenhorrn H, Muyzer G, van Hannen EJ, Gons HJ, Laanbroek HJ (1998) Divergent members of the bacterial division Verrucomicrobiales in a temperate freshwater lake. FEMS Microbiol Ecol 25:159–169
Acknowledgments
We thank Sidinei M. Thomas and the Universidade Estadual de Maringá, Núcleo de Pesquisas em Limnologia, Ictiologia e Aqüicultura, Maringá—PR—Brazil. We also thank A. M. Lemke, J. Bartletti, K. Kiehl, and A. K. da Silva Nakamura for field assistance, S. Paver for improvements to the manuscript, and boatmen A. Soares and Roberto. Brazil samples were collected under CNPq permit Portaria MCT no. 260/03, and work was partially supported by CNPq (Long Term Ecological Program, Brazilian site 6). Work at the American Museum of Natural History was supported by the Lewis and Dorothy Cullman Program in Molecular Systematics, with funding from the Sackler Institute of Comparative Genomics. E. K. Lienau and J. Rothe acknowledge financial assistance from the MacCracken Fellowship (NYU) and Hudson River Foundation, NY, respectively.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM
(PDF 23.6 kb)
Rights and permissions
About this article
Cite this article
Lemke, M.J., Lienau, E.K., Rothe, J. et al. Description of Freshwater Bacterial Assemblages from the Upper Paraná River Floodpulse System, Brazil. Microb Ecol 57, 94–103 (2009). https://doi.org/10.1007/s00248-008-9398-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-008-9398-3