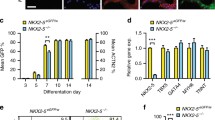
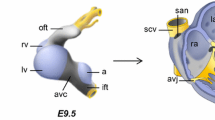
Abstract
Cell specification and differentiation of cardiomyocytes from mesodermal precursors is orchestrated by epigenetic and transcriptional inputs throughout heart formation. Of the many transcription factor super families that play a role in this process, the basic Helix-loop Helix (bHLH) family of proteins is well represented. The bHLH protein by design allows for dimerization—both as homodimers and heterodimers with other proteins within the family. Although DNA binding is mediated via a short variable cis-element termed an E-box, it is clear that DNA-affinity for these elements as well as the transcriptional input conveyed is dictated largely by the transcriptional partners within the dimer complex. Dimer partner choice has a number of inputs requiring co-expression within a given cell nucleus and dimerization modulation by the level of protein present, and post-translational modifications that can both enhance or reduce protein–protein interactions. Due to these complex interrelationships, it has been difficult to identity bona-fide downstream transcriptional targets and define the molecular pathways regulated of bHLH factors within cardiogenesis, despite the clear roles suggested via loss-of-function animals models. This review focuses on the Hand bHLH proteins—key members of the Twist-family of bHLH factors. Despite over a decade of investigation, questions regarding functional redundancy, downstream targets, and biological role during heart specification and differentiation have still not been fully addressed. Our goal is to review what is currently known and address strategies for gaining further understanding of Hand/Twist gene dosage and functional redundancy relationships within the developing heart that may underlie congenital heart defect pathogenesis.



Similar content being viewed by others
References
Barbosa AC, Funato N, Chapman S, McKee MD, Richardson JA, Olson EN, Yanagisawa H (2007) Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev Bio 310:154–168
Barnes RM, Firulli AB (2009) A Twist of insight, the role of Twist-Family bHLH factors in development. Int J Dev Biol 53(7):909–924
Breckenridge R, Zuberi Z, Gomes J, Orford R, Dupays L, Felkin LE, Clark JE, Magee AI, Ehler E, Birks EJ et al (2009) Overexpression of the transcription factor Hand1 causes predisposition towards arrhythmia in mice. J Molec Cell Cardiol 47:133–141
Castanon I, Von Stetina S, Kass J, Baylies MK (2001) Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development 128:3145–3159
Chen ZF, Behringer RR (1995) Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev 9:686–699
Connerney J, Andreeva V, Leshem Y, Muentener C, Mercado MA, Spicer DB (2006) Twist1 dimer selection regulates cranial suture patterning and fusion. Dev Dyn 235:1345–1357
Connerney J, Andreeva V, Leshem Y, Mercado MA, Dowell K, Yang X, Lindner V, Friesel RE, Spicer DB (2008) Twist1 homodimers enhance FGF responsiveness of the cranial sutures and promote suture closure. Dev Biol 318:323–334
Cross JC, Flannery ML, Blanar MA, Steingrimsson E, Jenkins NA, Copeland NG, Rutter WJ, Werb Z (1995) Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development 121:2513–2523
Cserjesi P, Brown D, Lyons GE, Olson EN (1995) Expression of the novel basic helix-loop-helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev Biol 170:664–678
Dai YS, Cserjesi P, Markham BE, Molkentin JD (2002) The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J Biol Chem 277:24390–24398
Fernandez-Teran M, Piedra ME, Kathiriya IS, Srivastava D, Rodriguez-Rey JC, Ros MA (2000) Role of dHAND in the anterior-posterior polarization of the limb bud: implications for the sonic hedgehog pathway. Development 127:2133–2142
Firulli AB (2003) A HANDful of questions: the molecular biology of the HAND-subclass of basic helix-loop-helix transcription factors. Gene 312C:27–40
Firulli AB, Conway SJ (2004) Combinatorial transcriptional interaction within the cardiac neural crest: a pair of HANDs in heart formation. Birth Defects Res C Embryo Today 72:151–161
Firulli AB, Thattaliyath BD (2002) Transcription factors in cardiogenesis: the combinations that may unlock the mysteries of the heart. Int Rev Cytol 214:1–62
Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN (1998) Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet 18:266–270
Firulli BA, Hadzic DB, McDaid JR, Firulli AB (2000) The basic helix-loop-helix transcription factors dHAND and eHAND exhibit dimerization characteristics that suggest complex regulation of function. J Biol Chem 275:33567–33573
Firulli B, Howard MJ, McDaid JR, McIlreavey L, Dionne KM, Centonze V, Cserjesi P, Virshup DMa, Firulli AB (2003) PKA, PKC and the protein phosphatase 2A influence HAND factor function: a mechanism for tissue specific transcriptional regulation. Mol Cell 12:1225–1237
Firulli BA, Krawchuk D, Centonze VE, Virshup DE, Conway SJ, Cserjesi P, Laufer E, Firulli AB (2005) Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet 37:373–381
Firulli BA, Redick BA, Conway SJ, Firulli AB (2007) Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J Biol Chem 282:27536–27546
Heidt AB, Rojas A, Harris IS, Black BL (2007) Determinants of myogenic specificity within MyoD are required for noncanonical E box binding. Mol Cell Biol 27:5910–5920
Hill AA, Riley PR (2004) Differential regulation of Hand1 homodimer and Hand1-E12 heterodimer activity by the cofactor FHL2. Mol Cell Biol 24:9835–9847
Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H (1995) Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol 15:3813–3822
Jabs EW (2004) TWIST and Saethre-Chotzen syndrome. In: Epstein CJ, Erickson RP, Wynshaw-Boris A (eds) Inborn errors of development. Oxford University Press, New York, pp 401–409
Knofler M, Meinhardt G, Bauer S, Loregger T, Vasicek R, Bloor DJ, Kimber SJ, Husslein P (2002) Human Hand1 basic helix-loop-helix (bHLH) protein: extra-embryonic expression pattern, interaction partners and identification of its transcriptional repressor domains. Biochem J 361:641–651
Lo PCH, Zaffran S, Senatore S, Frasch M (2007) The drosophila hand gene is required for remodeling of the developing adult heart and midgut during metamorphosis. Dev Bio 311:287–296
Martindill DMJ, Risebro CA, Smart N, Franco-Viseras MDM, Rosario CO, Swallow CJ, Dennis JW, Riley PR (2007) Nucleolar release of Hand1 acts as a molecular switch to determine cell fate. Nat Cell Biol 9:1131–1141 (see comment)
Massari ME, Murre C (2000) Helix-Loop-Helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20:429–440
McFadden DG, McAnally J, Richardson JA, Charité J, Olson EN (2002) Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development 129:3077–3088
McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN (2005) The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132:189–201
Morikawa Y, Cserjesi P (2004) Extra-embryonic vasculature development is regulated by the transcription factor HAND1. Development 131:2195–2204
Morikawa Y, Cserjesi P (2008) Cardiac neural crest expression of Hand2 regulates outflow and second heart field development. Circ Res 103:1422–1429
Nemer G, Fadlalah F, Usta J, Nemer M, Dbaibo G, Obeid M, Bitar F (2006) A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Hum Mutat 27:293–294
Olson EN (1993) Regulation of muscle transcription by the MyoD family. The heart of the matter. Circ Res 72:1–6
Olson EN (1993) Signal transduction pathways that regulate skeletal muscle gene expression. Mol Endocrinol 7:1369–1378
Olson EN (2002) A genetic blueprint for growth and development of the heart. Harvey Lect 98:41–64
Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, Evans SM, Clark B, Feramisco JR, Giles W, Ho SY, Benson DW, Silberbach M, Shou W, Chien KR (2004) Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117:373–386
Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, Van Rheeden R, Wilson DB, Watson MS, Hing AV (1999) GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am J Med Genet 83:201–206
Peters TJ, Chapman BM, Soares MJ (2000) Trophoblast differentiation: an in vitro model for trophoblast giant cell development. In: Tuan RS, Lo CW (eds) Developmental biology protocols. Humana Press Inc., Totowa, pp 301–311
Reamon-Buettner SM, Ciribilli Y, Inga A, Borlak J (2008) A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Hum Mol Genet 17:1397–1405
Riley P, Anson-Cartwright L, Cross JC (1998) The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet 18:271–275
Risebro CA, Smart N, Dupays L, Breckenridge R, Mohun TJ, Riley PR (2006) Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development 133:4595–4606
Sakabe M, Matsui H, Sakata H, Ando K, Yamagishi T, Nakajima Y (2005) Understanding heart development and congenital heart defects through developmental biology: a segmental approach. Congenit Anom 45:107–118
Srivastava D, Cserjesi P, Olson EN (1995) A subclass of bHLH proteins required for cardiac morphogenesis. Science 270:1995–1999
Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN (1997) Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet 16:154–160
Thattaliyath BD, Firulli BA, Firulli AB (2002) The Basic-Helix-Loop-Helix transcription factor HAND2 directly regulates transcription of the atrial naturetic peptide gene. J Mol Cell Cardiol 34:1325–1344
Togi K, Kawamoto T, Yamauchi R, Yoshida Y, Kita T, Tanaka M (2004) Role of Hand1/eHAND in the dorso-ventral patterning and interventricular septum formation in the embryonic heart. Mol Cell Biol 24:4627–4635
Vincentz JW, Barnes RM, Rodgers R, Firulli BA, Conway SJ, Firulli AB (2008) An Absence of Twist1 results in aberrant cardiac neural crest morphogenesis. Dev Biol 320:131–139
Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DYR (2000) The bHLH transcription factor HAND2 plays parallel roles in zebrafish heart and pectoral fin development. Development 127:2573–2582
Acknowledgments
We would like to thank members of the Firulli and Conway labs for scientific assistance. We would also like to thank the Riley Heart Research Center for helpful input during robust group discussions. Support is provided by the Herman B Wells Center for Pediatric Research, the Riley Children’s Foundation, and the Division of Pediatric Cardiology and by NIH P01HL085098 (ABF & SJC).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Conway, S.J., Firulli, B. & Firulli, A.B. A bHLH Code for Cardiac Morphogenesis. Pediatr Cardiol 31, 318–324 (2010). https://doi.org/10.1007/s00246-009-9608-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00246-009-9608-x