Abstract
We propose an extension of the distance matrix methods NJst and ASTRID to infer species trees from incongruent gene trees having Incomplete Lineage Sorting. Both approaches consider the average internode distance (ID) between individual taxa pairs as the distance measure. The measure ID does not use the root of a tree, and thus may not always infer the relative position of a taxon with respect to the root. We define a novel distance measure excess gene leaf count (XL) between individual couplets. The XL measure is computed using the root of a tree. It is proved to be additive, and is shown to infer the relative order of divergence among individual couplets better. We propose a novel method IDXL which uses both the XL and ID measures for species tree construction. IDXL is shown to perform better than NJst and other distance matrix approaches for most of the biological and simulated datasets. Having the same computational complexity as NJst, IDXL can be applied for species tree inference on large-scale biological datasets.















Similar content being viewed by others
Notes
Statistical consistency means that as the number of error-free gene trees approaches infinity, the method recovers the correct species tree with high probability.
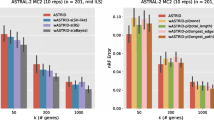
details of this dataset is discussed in “Experimental Results” section.
Here K denotes \(10^3,\) and M denotes \(10^6.\)
Values inside the third bracket and separated by ‘/,’ denote multiple model conditions.
References
Ané C, Larget BR, Baum DA, Smith SD, Rokas A (2007) Bayesian estimation of concordance among gene trees. Mol Biol Evol 24(2):412–426
Baum DA (2007) Concordance trees, concordance factors, and the exploration of reticulate genealogy. Taxon 56(2):417–426
Bayzid MS, Warnow T (2012) Estimating optimal species trees from incomplete gene trees under deep coalescence. J Comput Biol 19(6):591–605
Bayzid MS, Warnow T (2013) Naive binning improves phylogenomic analyses. Bioinformatics 19:1–16. doi:10.1093/bioinformatics/btt394
Bayzid MS, Hunt T, Warnow T (2014) Disk covering methods improve phylogenomic analyses. BMC Genomics 15(Suppl 6, S7):1–11. doi:10.1186/1471-2164-15-S6-S7
Bhattacharyya S, Mukhopadhyay J (2016) Accumulated coalescence rank and excess gene count for species tree inference. In: AlCOB. LNBI, vol 9096. Springer, Cham, pp 93–105
Bogdanowicz D, Giaro K, Wröbel B (2012) TreeCmp: comparison of trees in polynomial time. Evol Bioinform 8:475–487
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard M, Rambaut A, Drummond AJ (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10(4):1–6. doi:10.1371/journal.pcbi.1003537
Bryant D, Steel M (2009) Computing the distribution of a tree metric. IEEE/ACM Trans Comput Biol Bioinform 6(3):420–426
Buneman P (1974) A note on the metric properties of trees. J Combin Theory Ser B 17(1):48–50
Carstens BC, Knowles LL (2007) Estimating species phylogeny from gene-tree probabilities despite incomplete lineage sorting: an example from melanoplus grasshoppers. Syst Biol 56(3):400–411
Chaudhary R, Bansal MS, Wehe A, Fernández-Baca D, Eulenstein O (2010) iGTP: a software package for large-scale gene tree parsimony analysis. BMC Bioinform 23(574):1–7
Chaudhary R, Burleigh JG, Fernández-Baca D (2013) Inferring species trees from incongruent multi-copy gene trees using the Robinson-Foulds distance. Algorithms Mol Biol 8(28):1–12
Chaudhary R, Burleigh JG, Fernández-Baca D (2015) MulRF: a software package for phylogenetic analysis using multi-copy gene trees. Bioinformatics 31(3):432–433
Chiari Y, Cahais V, Galtier N, Delsuc F (2012) Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria). BMC Biol 10(65):1–14
Chifman J, Kubatko L (2014) Quartet Inference from SNP data under the coalescent model. Bioinformatics 30(23):3317–3324
Chifman J, Kubatko L (2015) Identifiability of the unrooted species tree topology under the coalescent model with time-reversible substitution processes, site-specific rate variation, and invariable sites. J Theor Biol 374:35–47
Chou J, Gupta A, Yaduvanshi S, Davidson R, Nute M, Mirarab S, Warnow T (2015) A comparative study of SVDQuartets and other coalescent-based species tree estimation methods. BMC Genomics 16(Suppl 10, S2):1–11. doi:10.1186/1471-2164-16-S10-S2
Dasarathy G, Nowak R, Roch S (2015) Data requirement for phylogenetic inference from multiple loci: a new distance method. IEEE/ACM Trans Comput Biol Bioinform 12(2):422–432
DeGiorgio M, Degnan JH (2010) Fast and consistent estimation of species trees using supermatrix rooted triples. Mol Biol Evol 27(3):552–569
DeGiorgio M, Degnan J (2014) Robustness to divergence time underestimation when inferring species trees from estimated gene trees. Syst Biol 63(1):66–82
Degnan JH, Rosenberg NA (2009) Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol 24(6):332–340
Deonier RC, Tavaré S, Waterman M (2005) Computational genome analysis: an introduction. Springer, New York. doi:10.1007/0-387-28807-4
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. Mol Biol Evol 7(214):1–8
Durand D, Halldorsson BV, Vernot B (2005) A hybrid micro-macroevolutionary approach to gene tree reconstruction. J Comput Biol 13(2):320–335
Edwards SV, Liu L, Pearl DK (2007) High-resolution species trees without concatenation. PNAS 104(14):5936–5941
Fan HH, Kubatko LS (2011) Estimating species trees using approximate Bayesian computation. Mol Phys Evol 59(2):354–363
Felsenstein J (2003) Inferring phylogenies. Sinauer Associates, Sunderland
Felsenstein J (2013) The Newick tree format. http://evolution.genetics.washington.edu/phylip/newicktree.html. Accessed 2 May 2013
Fletcher W, Yang Z (2009) INDELible: a flexible simulator of biological sequence evolution. Mol Biol Evol 26(8):1879–1888
Gascuel O (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14(7):685–695
Gascuel O (2000) Data model and classification by trees: the minimum variance reduction (MVR) method. J Classif 17(1):67–99
Hartmann K, Wong D, Stadler T (2010) Sampling trees from evolutionary models. Syst Biol 59(4):465–476
Heled J, Drummond AJ (2010) Bayesian inference of species trees from multilocus data. Mol Biol Evol 27(3):570–580
Helmkamp LJ, Jewett EM, Rosenberg NA (2012) Improvements to a class of distance matrix methods for inferring species trees from gene trees. J Comput Biol 19(6):632–649
Jewett EM, Rosenberg NA (2012) iGLASS: an improvement to the GLASS method for estimating species trees from gene trees. J Comput Biol 19(3):293–315
Jiang T, Kearney P, Li M (2001) A polynomial time approximation scheme for inferring evolutionary trees from quartet topologies and its application. SIAM J Comput 30(6):1942–1961
Jones NC, Pevzner PA (2004) An introduction to bioinformatics algorithms (computational molecular biology). MIT, Cambridge
Kingman JFC (1982) On the genealogy of large populations. J Appl Probab (Essays in Statistical Science) 19A:27–43
Kubatko LS, Degnan JH (2007) Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst Biol 56(1):17–24
Kubatko LS, Carstens BC, Knowles L (2009) STEM: species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics 25(7):971–973
Larget BR, Kotha SK, Dewey CN, Ané C (2010) BUCKy: gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics 26(22):2910–2911
Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25(7):1307–1320
Lin Y, Rajan V, Moret BME (2012) A metric for phylogenetic trees based on matching. IEEE/ACM Trans Comput Biol Bioinform 9(4):1014–1022
Liu K (2011) RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation. PLoS ONE 6(11):e27731. doi:10.1371/journal.pone.0027731
Liu L (2008) BEST: Bayesian estimation of species trees under the coalescent model. Bioinformatics 24(21):2542–2543
Liu L, Pearl DK (2007) Species trees from gene trees: reconstructing Bayesian posterior distributions of a species phylogeny using estimated gene tree distributions. Syst Biol 56(3):504–514
Liu L, Yu L (2011) Estimating species trees from unrooted gene trees. Syst Biol 60(5):661–667
Liu L, Pearl DK, Brumfield RT, Edwards SV (2008) Estimating species trees using multiple-allele DNA sequence data. Evolution 62(8):468–477
Liu L, Yu L, Pearl DK, Edwards SV (2009) Estimating species phylogenies using coalescence times among sequences. Syst Biol 58(5):468–477
Liu L, Yu L, Edwards SV (2010a) A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol Biol 10(302):1–18
Liu L, Yu L, Pearl DK (2010b) Maximum tree: a consistent estimator of the species tree. J Math Biol 60(1):95–106
Liu L, Xi Z, Davis CC (2015a) Coalescent methods are robust to the simultaneous effects of long branches and incomplete lineage sorting. Mol Biol Evol 32(3):791–805. doi:10.1093/molbev/msu331
Liu L, Xi Z, Wu S, Davis CC, Edwards SV (2015b) Estimating phylogenetic trees from genome-scale data. Ann N Y Acad Sci 1360(1):36–53. doi:10.1111/nyas.12747
Ma B, Li M, Zhang L (2000) From gene trees to species trees. SIAM J Comput 30(3):729–752
Maddison WP (1997) Gene trees in species trees. Syst Biol 46(3):523–536
Maddison WP, Knowles LL (2006) Inferring phylogeny despite incomplete lineage sorting. Syst Biol 55(1):21–30
Mailund T (2015) On gene trees and species trees. http://www.mailund.dk/index.php/2009/02/12/on-gene-trees-and-species-trees/. Accessed 27 June 2015
Mallo D, de Oliveira ML, Posada D (2015) SimPhy: phylogenomic simulation of gene, locus and species trees. Syst Biol 65(2):1–37. doi:10.1093/sysbio/syv082
Mirarab S, Warnow T (2015) ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31(12):i44–i52
Mirarab S, Bayzid MS, Boussau B, Warnow T (2014a) Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science 346(6215):1–9
Mirarab S, Bayzid MS, Warnow T (2014b) Evaluating summary methods for multilocus species tree estimation in the presence of incomplete lineage sorting. Syst Biol 65(3):366–380. doi:10.1093/sysbio/syu063
Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T (2014c) ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30(17):i541–i548
Mossel E, Roch S (2010) Incomplete lineage sorting: consistent phylogeny estimation from multiple loci. IEEE/ACM Trans Comput Biol Bioinform 7(1):166–171
Nakhleh L (2013) Computational approaches to species phylogeny inference and gene tree reconciliation. Trends Ecol Evol 28(12):719–728
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Bioinformatics 26(7):1641–1650
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5(3):1–10. doi:10.1371/journal.pone.0009490
Rannala B, Yang Z (2003) Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164(4):1645–1656
Robinson DR, Foulds LR (1981) Comparison of phylogenetic trees. Math Biosci 53(1–2):131–147
Roch S, Steel M (2015) Likelihood-based tree reconstruction on a concatenation of aligned sequence data sets can be statistically inconsistent. Theor Popul Biol 100:56–62
Roch S, Warnow T (2015) On the robustness to gene tree estimation error (or lack thereof) of coalescent-based species tree methods. Syst Biol 64(4):663–676
Rokas A, Williams B, King N, Carroll S (2003) Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425:798–804
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Song S, Liu L, Edwards SV, Wu S (2012) Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc Natl Acad Sci USA 109(37):14,942–14,947
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313
Steel M, Penny D (1993) Distributions of tree comparison metrics–some new results. Syst Biol 42(2):126–141
Studier JA, Keppler KL (1988) A note on the neighbor-joining algorithm of Saitou and Nei. Mol Biol Evol 5(6):729–731
Sukumaran J, Holder MT (2000) DendroPy: a Python library for phylogenetic computing. Bioinformatics 26(12):1569–1571
Than C, Nakhleh L (2009) Species tree inference by minimizing deep coalescences. PLoS Comput Biol 5(9):1–12
Than C, Ruths D, Nakhleh L (2008) PhyloNet: a software package for analyzing and reconstructing reticulate evolutionary relationships. BMC Bioinform 9(322):1–16
Vachaspati P, Warnow T (2015) ASTRID: accurate species trees from internode distances. BMC Genomics 16(Suppl 10, S3):1–18. doi:10.1186/1471-2164-16-S10-S3
Wickett NJ et al (2014) Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci USA 111(45):E4859–E4868. doi:10.1073/pnas.1323926111
Wu Y (2011) Coalescent-based species tree inference from gene tree topologies under incomplete lineage sorting by maximum likelihood. Evolution 66(3):763–775
Xi Z, Liu L, Rest JS, Davis CC (2014) Coalescent versus concatenation methods and the placement of Amborella as sister to water lilies. Syst Biol 63(6):919–932
Yang Z (2014) Molecular evolution a statistical approach, 1st edn. Oxford University Press, Oxford
Yu Y, Warnow T, Nakhleh L (2011) Algorithms for MDC-based multi-locus phylogeny inference: beyond rooted binary gene trees on single alleles. J Comput Biol 18(11):1543–1559
Yule GU (1925) A mathematical theory of evolution, based on the conclusions of Dr. J. C. Willis, F.R.S. Philos Trans R Soc B 213(402–410):21–87
Zimmermann T, Mirarab S, Warnow T (2014) BBCA: Improving the scalability of *BEAST using random binning. BMC Genomics 15 (Suppl 6, S11):1–9
Acknowledgements
The first author acknowledges Tata Consultancy Services (TCS) for providing the research scholarship. We acknowledge the anonymous reviewers for their insightful comments and suggestions towards improvement of this work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bhattacharyya, S., Mukherjee, J. IDXL: Species Tree Inference Using Internode Distance and Excess Gene Leaf Count. J Mol Evol 85, 57–78 (2017). https://doi.org/10.1007/s00239-017-9807-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-017-9807-7