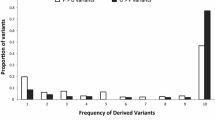
Abstract
Contrary to the classical view, a large amount of non-coding DNA seems to be selectively constrained in Drosophila and other species. Here, using Drosophila miranda BAC sequences and the Drosophila pseudoobscura genome sequence, we aligned coding and non-coding sequences between D. pseudoobscura and D. miranda, and investigated their patterns of evolution. We found two patterns that have previously been observed in comparisons between Drosophila melanogaster and its relatives. First, there is a negative correlation between intron divergence and intron length, suggesting that longer non-coding sequences may contain more regulatory elements than shorter sequences. Our other main finding is a negative correlation between the rate of non-synonymous substitutions (d N) and codon usage bias (F op), showing that fast-evolving genes have a lower codon usage bias, consistent with strong positive selection interfering with weak selection for codon usage.




Similar content being viewed by others
References
Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amantides PG et al (2000) The genome sequence of Drosophila melanogaster. Science 287:2185–2195
Akashi H (1995) Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139:1067–1076
Andolfatto P (2005) Adaptive evolution of non-coding DNA in Drosophila. Nature 437:1149–1152
Andolfatto P (2007) Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res 17:1755–1762
Arnone MI, Davidson EH (1997) The hardwiring of development: organization and function of genomic regulatory systems. Development 124:1851–1864
Babbitt GA, Kim Y (2008) Inferring natural selection on fine-scale chromatin organization in yeast. Mol Biol Evol 25(8):1714–1727
Bachtrog D (2007) Reduced selection for codon usage bias in Drosophila miranda. J Mol Evol 64:586–590
Bachtrog D (2008) Similar rates of protein adaptation in Drosophila miranda and D. melanogaster, two species with different current effective population sizes. BMC Evol Biol 8:334
Bachtrog D, Andolfatto P (2006) Selection, recombination and demographic history in Drosophila miranda. Genetics 174:2045–2059
Bachtrog D, Hom E, Wong KM, Maside X, de Jong P (2008) Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol 9:R30
Barrio E, Latorre A, Moya A, Ayala FJ (1992) Phylogenetic reconstruction of the Drosophila obscura group, on the basis of mitochondrial DNA. Mol Biol Evol 9:621–635
Bartolomé C, Charlesworth B (2006a) Rates and patterns of chromosomal evolution in Drosophila pseudoobscura and D. miranda. Genetics 173:779–791
Bartolomé C, Charlesworth B (2006b) Evolution of amino-acid sequences and codon usage on the Drosophila miranda neo-sex chromosomes. Genetics 174:2033–2044
Bartolomé C, Maside X, Yi S, Grant AL, Charlesworth B (2005) Patterns of selection on synonymous and nonsynonymous variants in Drosophila miranda. Genetics 169:1495–1507
Bergman CM, Kreitman M (2001) Analysis of conserved noncoding DNA in Drosophila reveals similar constraints in intergenic and intronic sequences. Genome Res 11:1335–1345
Bergman CM, Pfeiffer BD, Rincón-Limas DE, Hoskins RA, Gnirke A, Mungall CJ, Wang AM, Kronmiller B, Pacleb J, Park S, Stapleton M, Wan K, George RA, de Jong PJ, Botas J, Rubin GM, Celniker SE (2002) Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol 3(12):0086.1–0086.20
Betancourt AJ, Presgraves DC (2002) Linkage limits the power of natural selection in Drosophila. Proc Natl Acad Sci USA 99(21):13616–13620
Bierne N, Eyre-Walker A (2003) The problem of counting sites in the estimation of the synonymous and nonsynonymous substitution rates: Implications for the correlation between the synonymous substitution rate and codon usage bias. Genetics 165:1587–1597
Bierne N, Eyre-Walker A (2006) Variation in synonymous codon use and DNA polymorphism within the Drosophila genome. J Evol Biol 19:1–11
Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH et al (2007) Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature 447:799–816
Bradnam KR, Korf I (2008) Longer first introns are a general property of eukaryotic gene structure. PLoS ONE 3(8):e3093
Bray N, Pachter L (2004) MAVID: constrained ancestral alignment of multiple sequences. Genome Res 14:693–699
Casillas S, Barbadilla A, Bergman CM (2007) Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol Biol Evol 24(10):2222–2234
Charlesworth B (1994) The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet Res 63:213–227
Chen Y, Stephan W (2003) Compensatory evolution of a precursor messenger RNA secondary structure in the Drosophila melanogaster Adh gene. Proc Natl Acad Sci USA 100(20):11499–11504
Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA et al (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218
Comeron JM (1995) A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J Mol Evol 41:1152–1159
Comeron JM, Aguadé M (1996) Synonymous substitutions in the Xdh gene of Drosophila: Heterogeneous distribution along the coding region. Genetics 144:1053–1062
Comeron JM, Williford A, Kliman RM (2008) The Hill–Robertson effect: evolutionary consequences of weak selection and linkage in finite populations. Heredity 100:19–31
Drummond DA, Wilke CO (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134:341–352
Duret L (2001) Why do genes have introns? Recombination might add a new piece to the puzzle. Trends Genet 17(4):172–175
Duret L, Mouchiroud D (1999) Expression pattern and, surprisingly, gene length shape codon usage in Cænorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci USA 96(8):4482–4487
Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30(1):207–210
Emberly E, Rajewsky N, Siggia ED (2003) Conservation of regulatory elements between two species of Drosophila. BMC Bioinformatics 4:57
FLYBASE: a database of the Drosophila genome. http://www.flybase.org
Haddrill PR, Thornton KR, Charlesworth B, Andolfatto P (2005) Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol 6:R67
Haddrill PR, Bachtrog D, Andolfatto P (2008) Positive and negative selection on noncoding DNA in Drosophila simulans. Mol Biol Evol 25(9):1825–1834
Halligan DL, Keightley PD (2006) Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res 16:875–884
Halligan DL, Eyre-Walker A, Andolfatto P, Keightley PD (2004) Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res 14:273–279
Hardison RC (2000) Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet 16(9):369–372
Hill WG, Robertson A (1966) The effect of linkage on limits to artificial selection. Genet Res 8:269–294
Ikemura T (1981) Correlation between the abundance of E. coli transfer RNAs and the occurrence of the respective codon in the protein genes. J Mol Biol 146:1–21
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E (2009) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458:362–366
Keightley PD, Johnson T (2004) MCALIGN: stochastic alignment of noncoding DNA sequences based on an evolutionary model of sequence evolution. Genome Res 14:442–450
Kirby DA, Muse SV, Stephan W (1995) Maintenance of pre-mRNA secondary structure by epistatic selection. Proc Natl Acad Sci USA 92(20):9047–9051
Larracuente AM, Sackton TB, Greenberg AJ, Wong A, Singh ND, Sturgill D, Zhang Y, Oliver B, Clark AG (2008) Evolution of protein-coding genes in Drosophila. Trends Genet 24(3):114–123
Leicht BG, Muse SV, Hanczyc M, Clark AG (1995) Constraints on intron evolution in the gene encoding the myosin alkali light chain in Drosophila. Genetics 139:299–308
Majewski J, Ott J (2002) Distribution and characterization of regulatory elements in the human genome. Genome Res 12:1827–1836
Marais G, Mouchiroud D, Duret L (2001) Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. PNAS 98(10):5688–5692
Marais G, Mouchiroud D, Duret L (2003) Neutral effect of recombination on base composition in Drosophila. Genet Res 81:79–87
Marais G, Domazet-Lošo T, Tautz D, Charlesworth B (2004) Correlated evolution of synonymous and nonsynonymous sites in Drosophila. J Mol Evol 59:771–779
Marais G, Nouvellet P, Keightley PD, Charlesworth B (2005) Intron size and exon evolution in Drosophila. Genetics 170:481–485
Maroni G (1994) The organization of Drosophila genes. DNA Seq 4(6):347–354
Moriyama EN, Powell JR (1998) Gene length and codon usage bias in Drosophila melanogaster, Saccharomyces cerevisiae and Escherichia coli. Nucl Ac Res 23(13):3188–3193
Mount SM, Burks C, Hertz G, Stormo GD, White O, Fields C (1992) Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucl Ac Res 20(16):4255–4262
Parsch J (2003) Selective constraints on intron evolution in Drosophila. Genetics 165:1843–1851
Parsch J (2004) Functional analysis of Drosophila melanogaster gene regulatory sequences by transgene coplacement. Genetics 168:559–561
Petrov DA (2002) DNA loss and evolution of genome size in Drosophila. Genetica 115:81–91
Pollard DA, Bergman CM, Stoye J, Celniker SE, Eisen MB (2004) Benchmarking tools for the alignment of functional. BMC Bioinformatics 5:6
Reenan RA (2005) Molecular determinants and guided evolution of species-specific RNA editing. Nature 434:409–413
Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, Couronne O, Hua S, Smith MA, Zhang P, Liu J, Bussemaker HJ, van Batenburg MF, Howells SL, Scherer SE, Sodergren E, Matthews BB, Crosby MA, Schroeder AJ, Ortiz-Barrientos D, Rives CM, Metzker ML, Muzny DM, Scott G, Steffen D, Wheeler DA, Worley KC, Havlak P, Durbin KJ, Egan A, Gill R, Hume J, Morgan MB, Miner G, Hamilton C, Huang Y, Waldron L, Verduzco D, Clerc-Blankenburg KP, Dubchak I, Noor MA, Anderson W, White KP, Clark AG, Schaeffer SW, Gelbart W, Weinstock GM, Gibbs RA (2005) Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res 15:1–18
Rogic S, Montpetit B, Hoos HH, Mackworth AK, Ouellette BFF, Hieter P (2008) Correlation between the secondary structure of pre-mRNA introns and the efficiency of splicing in Saccharomyces cerevisiae. BMC Genomics 9:355
Shields DC, Sharp PM, Higgins DG, Wright F (1988) “Silent” sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol 5(6):704–716
Singh ND, Davis JC, Petrov DA (2005) X-linked genes evolve higher codon bias in Drosophila and Caenorhabditis. Genetics 171:145–155
Sironi M, Menozzi G, Comi GP, Cagliani R, Bresolin N, Pozzoli U (2005) Analysis of intronic conserved elements indicates that functional complexity might represent a major source of negative selection on non-coding sequences. Hum Mol Genet 14(17):2533–2546
Subramanian S, Kumar S (2004) Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics 168:373–381
Thornton K (2003) Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics 19(17):2325–2327
Vicario S, Moriyama EN, Powell JR (2007) Codon usage in twelve species of Drosophila. BMC Evol Biol 7:226
Vicoso B, Haddrill PR, Charlesworth B (2008) A multispecies approach for comparing sequence evolution of X-linked and autosomal sites in Drosophila. Genet Res 90:421–431
Wang J, Keightley PD, Johnson T (2006) MCALIGN2: Faster, accurate global pairwise alignment of non-coding DNA sequences based on explicit models of indel evolution. BMC Bioinformatics 7:292
Warnecke T, Batada NN, Hurst LD (2008) The impact of the nucleosome code on protein-coding sequence evolution in yeast. PLoS Genetics 4(11):e1000250
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13(5):555–556
Yi S, Charlesworth B (2000) Contrasting patterns of molecular evolution of the genes on the new and old sex chromosomes of Drosophila miranda. Mol Biol Evol 17(5):703–717
Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B (2007) Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature 450(7167):233–238
Acknowledgments
This study was conducted as part of the GENACT Project, funded by the Marie Curie Host Fellowships for Early Stage training to SMP, as part of the 6th Framework Programme of the European Commission. Brian Charlesworth was supported by the Royal Society. BAC sequencing and Daniel L. Halligan were funded by a Wellcome Trust and BBSRC grant to Peter D. Keightley and Brian Charlesworth. We thank Dr. Mark Dorris for isolating BACs and Dr. Jane Rogers at the Wellcome Trust Sanger Institute for organizing their sequencing. We also thank two anonymous reviewers for helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Marion de Procé, S., Halligan, D.L., Keightley, P.D. et al. Patterns of DNA-Sequence Divergence Between Drosophila miranda and D. pseudoobscura . J Mol Evol 69, 601–611 (2009). https://doi.org/10.1007/s00239-009-9298-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-009-9298-2