Abstract
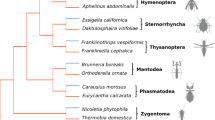
The Pten gene was initially identified in humans as a tumor suppressor. It has since been shown to play important roles in the control of cell size, cell motility, apoptosis, and organ size, and it has also been implicated in aging. Pten is highly conserved among organisms as diverse as nematodes, insects, and vertebrates. In contrast, a phylogenetic analysis by maximum likelihood of a 133-amino acid region showed an average nonsynonymous-to-synonymous rate ratio of 10.4 for Pten in the lineage leading to parasitoid wasps of the Nasonia genus, indicating very strong positive selection. A previous study identified Pten as a potential QTL candidate gene for differences in male wing size in Nasonia. Most of the amino acid replacements that occurred in the Nasonia lineage cluster in a small region of the protein surface, suggesting that they might be involved in an interaction between Pten and another protein. The phenotypic changes due to Pten are not yet known, although it is not associated with known differences in male wing size. Introgression of Pten from one species to another does affect longevity, but a causal relationship is not established.



Similar content being viewed by others
References
Anisimova M, Bielawski JP, Yang Z (2002) Accuracy and power of bayes prediction of amino acid sites under positive selection. Mol Biol Evol 19:950–958
Anisimova M, Nielsen R, Yang Z (2003) Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164:1229–1236
Bishop JG, et al. (2000) Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc Natl Acad Sci US 97:5322–5327
Bringold F, Serrano M (2000) Tumor suppressors and oncogenes in cellular senescence. Exp Gerontol 35:317–329
Coelho C, Leevers S (2000) Do growth and cell division rates determine cell size in multicellular organisms? J Cell Sci 113:2927–2934
Comeron JM (1995) A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J Mol Evol 41:1152–1159
Das S, Dixon JE, Cho W (2003) Membrane-binding and activation mechanism of PTEN. Proc Natl Acad Sci USA 100:7491–7496
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Ford MJ (2001) Molecular evolution of transferrin: evidence for positive selection in salmonids. Mol Biol Evol 18:639–647
Gadau J, Page R, Werren J (2002) The genetic basis of the interspecific differences in wing size in Nasonia (Hymenoptera; Pteromalidae): major quantitative trait loci and epistasis. Genetics 161:673–684
Goberdhan DC, Paricio N, Goodman EC, Mlodzik M, Wilson C (1999) Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev 13:3244–258
Goldberg A, Wildman DE, Schmidt TR, Huttemann M, Goodman M, Weiss ML, Grossman LI (2003) Adaptive evolution of cytochrome c oxidase subunit VIII in anthropoid primates. Proc Natl Acad Sci USA 100:5873–5878
Goldman N, Yang Z (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol 11:725–736
Hasegawa M, Iida Y, Yano T, Takaiwa F, Iwabuchi M (1985) Phylogenetic relationships among eukaryotic kingdoms inferred from ribosomal RNA sequences. J Mol Evol 22:32–38
Howard DJ (1999) Conspecific sperm and pollen precedence and speciation. Annu Rev Ecol Syst 30:109–132
Huelsenbeck JP, Rannala B (1997) Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science 276:227–232
Huttley GA, Easteal S, Southey MC, Tesoriero A, Giles GG, McCredie MR, Hopper JL, Venter DJ (2000) Adaptive evolution of the tumour suppressor BRCA1 in humans and chimpanzees. Australian Breast Cancer Family Study. Nat Genet 25:410–413
Hwangbo DS, Gersham B, Tu MP, Palmer M, Tatar M (2004) Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429:562–566
Jiggins FM, Hurst GDD, Yang Z (2002) Host-symbiont conflicts: positive selection on an outer membrane protein of parasitic but not mutualistic Rickettsiaceae. Mol Biol Evol 19:1341–1349
Kozma SC, Thomas G (2002) Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. BioEssays 24:65–71
Kumar S, Hedges SB (1998) A molecular timescale for vertebrate evolution. Nature 392:917–920
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163
Lee J, Yang H, Georgescu M, Di Cristofano A, Maehama T, Shi Y, Dixon J, Pandolfi P, Pavletich N (1999) Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 99:323–334
Li WH (1993) Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol 36:96–99
Li WH (1997) Molecular evolution. Sinauer Associates, Sunderland, MA
Maehama T, Dixon JE (1999) PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol 9:125–128
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Nielsen R, Yang Z (1998) Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148:929–936
Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, J Sninsky J, Adams MD, Cargill M (2005) A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol 3:e170
Pamilo P, Bianchi NO (1993) Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol Biol Evol 10:271–281
Podsypanina K, Hedrick Ellenson L, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R (1999) Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA 96:1563–1568
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Pupko T, Pe’er I, Shamir R, Graur D (2000) A fast algorithm for joint reconstruction of ancestral amino acid sequences. Mol Biol Evol 17: 890–896
Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV (1997) Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q233 that is mutated in multiple advanced cancers. Nat Genet 15:356–362
Suzuki Y, Gojobori T (1999) A method for detecting positive selection at single amino acid sites. Mol Biol Evol 16:1315–1328
Suzuki Y, Nei M (2004) False-positive selection identified by ML-based methods: examples from the Sig1 gene of the diatom Thalassiosira weissflogii and the tax gene of a human T-cell lymphotropic virus. Mol Biol Evol 21:914–921
Swanson WJ, Vacquier VD (2002) The rapid evolution of reproductive proteins. Nature Rev Gen 3:137–144
Swofford DL (2002) PAUP* Phylogenetic Analysis Using Parsimony (*and other methods). Sinauer Associates, Sunderland, MA
Tamura M, Gu J, Matsumoto K, Aota SI, Parsons R, Yamada KM (1998) Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 280:1614–1617
Urwin R, Holmes EC, Fox AJ, Derrick JP, Maiden MCJ (2002) Phylogenetic evidence for frequent positive selection and recombination in the meningococcal surface antigen PorB. Mol Biol Evol 19:1686–1694
Wallis M (2001) Episodic evolution of protein hormones in mammals. J Mol Evol 53:10–18
Ward T, Honeycutt R, Derr J (1997) Nucleotide sequence evolution at the kappa-casein locus: evidence for positive selection within the family Bovidae. Genetics 147:1863–1872
Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562
Weng LP, Brown JL, Baker KM, Ostrowski MC, Eng C (2002) PTEN blocks insulin-mediated ETS-2 phosphorylation through MAP kinase, independently of the phosphoinositide 3-kinase pathway. Hum Mol Genet 11:1687–1696
Weston R, Qureshi I, Werren JH (1999) Genetics of a morphological difference between two insect species. J Evol Biol 12:586–595
Wong WS, Yang Z, Goldman N, Nielsen R (2004) Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168:1041–1051
Yamada KM, Araki M (2001) Tumor suppressor PTEN: modulator of cell signaling, growth, migration and apoptosis. J Cell Sci 114:2375–2382
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yang Z (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15:568–573
Yang Z (2000) Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza virus. A J Mol Evol 51:423–432
Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. TREE 15:496–503
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Yang Z, Swanson WJ, Vacquier VD (2000) Maximum-likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol Biol Evol 17:1446–1455
Zhang J (2004) Frequent false detection of positive selection by the likelihood method with branch-site models. Mol Biol Evol 21:1332–1339
Zhang J, Rosenberg HF (2002) Diversifying selection of the tumor-growth promoter angiogenin in primate evolution. Mol Biol Evol 19:438–445
Zhang J, Rosenberg HF, Nei M (1998) Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci USA 95:3708–3713
Acknowledgments
We thank F. Depaulis, C. Terzian, and two anonymous reviewers for valuable comments on the manuscript and L. Enders and J. Traggis for assistance. This research was supported by grants from the U.S. National Science Foundation (DEB 9981637) and a seed grant to J.H.W. from the University of Rochester Nathan Shock Center (NIH P30AG18254).
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Willie J. Swanson]
Rights and permissions
About this article
Cite this article
Baudry, E., Desmadril, M. & Werren, J. Rapid Adaptive Evolution of the Tumor Suppressor Gene Pten in an Insect Lineage. J Mol Evol 62, 738–744 (2006). https://doi.org/10.1007/s00239-005-0002-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-005-0002-x