Abstract
Human monkeypox has attracted attention recently. Monkeypox virus (MPXV) keeps evolving as it spreading around the world rapidly, which may threaten the health of more and more people. Here, we have developed a high order reference method based on digital PCR (dPCR) for MPXV detection, of which the limits of quantification (LoQ) and detection (LoD) are 38 and 6 copies/reaction, respectively. Pseudovirus reference materials (RM) containing the conserved F3L gene has been developed, and the homogeneity assessment showed that the RM was homogeneous. The reference value with its expanded uncertainty determined by the established dPCR is (2.74 ± 0.46) × 103 copies/μL. Six different MPXV test kits were accessed by the RM. Four out of six test kits cannot reach their claimed LoDs. The poor analytical sensitivity might cause false-negative results, which lead to incorrect diagnosis and treatment. The establishment of a high order reference method of dPCR and pseudovirus RM is very useful for improving the accuracy and reliability of MPXV detection.
Graphical abstract

Similar content being viewed by others
Introduction
Since the first human case infected with monkeypox was discovered in 1970, its natural host reservoir keeps unknown, though plenty of mammalian species can be infected. Major of the human monkeypox cases were reported from Central and West Africa during past 70 years [1, 2]. The first infected case outside Africa was detected in the USA in 2003 [3] Surprisingly, monkeypox received worldwide attention again in recent years. Up to now, 78 countries and regions have reported monkeypox cases, totaling more than 60,000 cases (https://extranet.who.int/publicemergency). There have been 55 death cases since January 1, 2022, as reported by centers for Disease Control and Prevention in the USA (https://www.cdc.gov/poxvirus/monkeypox/response/2022/world-map.html). The higher fatality rate was found in higher in young people and those with immune deficiency [4].
Human monkeypox virus (MPXV) is member of the family Poxviridae, genus Orthopoxvirus. It was characterized into “high threat” biodefence category in the EU [5]. Its biosafety threat may increase with continuous spreading and virus evolution leading to increased worldwide health risk [6]. Analysis of genome sequencing showed that 2022 MPXV segregated in a divergent phylogenetic branch, reflecting accelerated evolution rate [7]. To deal with emerging monkeypox breakouts, it is important to ensure accuracy of diagnosis. Detection methods based on PCR technology and sequencing are less time-consuming than viral culture [8, 9]. Former research had established real time PCR (qPCR) assays for the specific detection of West African and Congo Basin MPXV DNA, which showed good sensitivity and rapidity [10]. However, the quantification result of qPCR assays relies on the standard curve [11]. NIST has just released their DNA control material for validation MPVX tests to support the urgent public health need as quickly as possible without compromising on quality (https://www.nist.gov/news-events/news/2022/07/nist-develops-genetic-material-validating-monkeypox-tests). There is still lack of high order reference method for MPXV quantification, as well as Pseudovirus reference material (RM) to provide quality control for the whole detection process.
Digital PCR technology can provide absolute quantitative result with a better comparability among different laboratories and platforms [12, 13]. The aim of this paper are to (1) establish high order reference method based on dPCR technology for quantification of MPXV, (2) develop a Pseudovirus RM for validation of the current method and kits, and (3) evaluate the performance of six commercial testing kits using the Pseudovirus RM.
Materials and methods
Preparation of Pseudovirus reference material
MPXV F3L gene was synthesized in vitro (Sangon Biotech, Shanghai, China). The gene sequences were cloned into replication deficient adenovirus vector (Ad5 type) [14]. The adenovirus vector containing the target gene was dissolved in 500 μL DMEM liquid medium, which was shaken and mixed well. 293A cells were inoculated in a 6-well plate and cultured in a cell incubator (37 ℃, 5%CO2) until the cells covered 80% of the plate. Then the plasmids were transfected into 293A cells (Sangon Biotech, Shanghai, China). After 48 h culture, the cell lysate were collected and purified by density gradient centrifugation at high speed. Pseudovirus were diluted to a certain concentration and packed into 1.5-ml cryopreservation tubes, 200 μL for each tube. The candidate Pseudovirus RMs were stored at − 80℃ for further use.
DNA extraction and purification
The Pseudovirus DNA was extracted and purified by using the DNeasy Blood & Tissue Kit (QIAGEN, Cat. 69,506) according the instruction. The purified DNA was accessed by measuring the absorbance at A260, A280, and A230 using NanoDrop 2000 (Thermo Fisher, USA).
dPCR method
The sequence information of MPXV was obtained from the National Biotechnology Information Center (National Center for Biotechnology Information, NCBI). The corresponding primers and probe were designed targeting F3L gene (NCBI Gene ID: 928,998), and the relevant literature was consulted and compared with SnapGene Viewer software. The specific sequences of primers and probe designed for MPXV F3L genes were shown in Table 1.
The dPCR amplification procedure was for 10 min at 94 °C, then for 30 s at 94 °C, for 1 min at 57 °C, 40 Cycles, and for 5 min at 68 °C. In order to ensure the integrity of water-in-oil droplets during the reaction, the temperature rising and cooling rate in the dPCR reaction program was set to 2 ℃/s.
Homogeneity and stability assessment
Eeleven vials of Pseudovirus RM were randomly selected and extracted for homogeneity analysis according to ISO Guide 35. Each vial was measured with three replicates by dPCR. F test was used to assess the homogeneity. The RM was stored at −80 ℃ for a long term. For stability analysis, three vials were extracted and measured with three replicates by dPCR at month 1, 2, 3, and 4. Trend analysis was used for the long-term stability according to ISO Guide 35 [15].
Characterization of RM and evaluation of uncertainty
Thirteen vials of Pseudovirus RM were randomly selected for value assignment. DNA was extracted and copy number concentration of F3L gene was determined by dPCR. Each extract was measured with three replicates. All data follows in a normal distribution tested by Shapiro–Wilk test. No outlier was found by Dixton and Grabbs’ test. The arithmetic mean of all measured data was calculated as the reference value of the Pseudovirus RM.
Uncertainty evaluation of the RM is mainly consisted of following factors: characterization (value assignment) (u1), DNA extraction efficiency (u2), homogeneity (u3), and stability in long-term storage (u4). Combined uncertainty (uc) was calculated using the equation below:
The expanded uncertainty (U) was calculated using the following equation, where the k is the coverage factor, taken as 2 at 95% confident level.
Results and discussion
Establishment of dPCR for MPXV detection
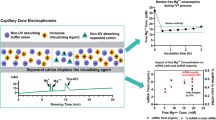
The annealing temperature was optimized firstly for the designed primers and probe. The thermal gradient was set by ranging amplification temperature from 54 to 58 ℃. Good separation between positive and negative fluorescence amplitude was observed for all annealing temperature, as shown in Fig. 1a. 57℃ was selected for the subsequent experiments as a better relative standard deviation (RSD), and higher copy number concentration was obtained. The primer and probe working concentration were optimized and fixed as 500 nM and 300 nM, respectively (Fig. 1b). Then, the dynamic range of dPCR was evaluated by using tenfold diluted samples containing F3L gene.
Establishment of dPCR method and reference material for detecting MPXV. The selection of annealing temperature was displayed in (a), dPCR amplification was set at the following annealing temperature gradients: 54℃, 55℃, 56℃, 57℃, and 58℃. The optimization of primer and probe concentrations were showed in (b), the different primer concentrations (300 nM, 400 nM, 500 nM, 600 nM) and probe concentrations (200 nM, 300 nM, 400 nM) were contained in orthogonal test. c The linearity range of the established dPCR method. The concentration of templates range from 38 to 9590 copies/reaction. d Homogeneity test result: eleven units were used for the homogeneity test. F tests was employed to statistically analyze the difference from intra and inter-units, and no significant distinction was observed between them. e Six qPCR kits were evaluated by the reference material. The dotted line represents the value of reference material quantified by dPCR
dPCR assay displayed an ideal linearity within the range of 4 orders of magnitude (R2 = 0.9997, slop is equal to 1.0141) (Fig. 1c). The slop and the correlation coefficient indicated the trueness was high as the measured concentration was very close to its corresponding nominal concentration by gravimetric dilution. Also, the precision of dPCR (represented by RSD) was less than 16% in the quantitative range, especially good (RSD <4%) for an optimum input concentration (Table S1 in supplementary), which indicated the precision is good.
The limit of detection (LoD) and quantification (LoQ) of dPCR were determined as 6 copies/reaction and 38 copies/reaction according to the criteria required by ISO17511 [16]. The LoD and LoQ was comparable with our previous report for the dPCR method for detection of SARS-CoV-2 [17]. With the high accuracy and good analytical sensitivity, it is suitable as a high order reference method for value assignment of RM.
Characterization of Pseudovirus RM
Compared with plasmid DNA RM, Pseudovirus RM can provide quality control for the the process of DNA extraction and detection. Therefore, the MPXV Pseudovirus RM was developed in this study. The homogeneity of the candidate Pseudovirus RM was accessed according to ISO Guide 35 [15]. The overall disperse was within 3.5% (RSD) shown in Fig. 1d. F test showed that it is homogeneous (Table S2 in the supplementary information).
Homogeneity and stability are the two major properties of a RM. For the long-term stability, not enough data was obtained currently as the RM was developed only for 4 months (Table S3 in the supplementary information). However, we have other similar Pseudovirus RM that can be stable for 6 months under −20 °C according to our previous data (https://www.ncrm.org.cn/Web/Ordering/MaterialDetail?autoID=25152). Thus, we speculate the Pseudovirus RM for MPXV developed in this study can be stable for at least 6 months under −80 °C. The RM was then characterized by the established dPCR method. The reference value and its expanded uncertainty were determined to be 2.74 × 103 and 0.46 × 103 copies/μL (Fig. 1d). The uncertainty was mainly consisted of those from the quantification method, homogeneity, and long-term storage (Table S4 in the supplementary information).
Evaluation of qPCR kits performance using the Pseudovirus RM
Six different commercial qPCR testing kits for F3L gene were evaluated using proposed Pseudovirus RM (Fig. 1e). The qPCR information was listed in Table S5 in the supplemental information. The RSD of quantitative results provided by six kits was less than 5%. Six standard curves were generated by quantifying the serial dilution of the reference material. The linear correlation coefficient (R2) of six standard curves was higher than 0.9920. However, the amplification efficiency (E) varied from 85 ~106%. Kit 4 showed amplification efficiency of 100%. The LoQs of these kits ranged from 18 to 82 copies/reaction, according to the criteria that relative standard deviation (RSD) less than 25% (Fig. 2a–g) [18].
Evaluation of qPCR kits performance using MPXV Pseudovirus reference material. Standard curves was obtained from serial-diluted reference material using six qPCR kits: a Kit-1; b Kit-2; c Kit-3; d Kit-4; e Kit-5; f Kit-6. For standard curves targeting F3L gene, the template concentrations ranged from 3 to 2080 copies/μL. Error bar represented the standard deviation of the mean. The arrow head pointed to the LoQ of each assay. The LoQs of six qPCR kits are summarized in (g). Reference material was diluted to the concentration of claimed LoDs
Lastly, we verified the LoDs claimed by the manufacturers. Surprisingly, only two kits (Kit3 and Kit 4) reached the given LoD among 20 repeats, while the LoD of other kits was inconsistent with the claimed ones. Especially, Kit 2 reported only two positive results among 20 repeats with the concentration of LoD (Fig. 2g). The inconsistence is probably caused by 3 factors: (1) the instability of qPCR system; (2) the different amplification efficiencies of reference materials (standard curve) and samples; and (3) the influence of potential inhibitors in matrix. It is necessary to know these effects before using the kit in clinical diagnosis. The poor analytical sensitivity might cause false-negative results, which lead to wrong diagnosis and treatment. To avoid misleading, it would be better to evaluate the performance of the commercial kits by validated RM before using.
As dPCR is independent of standard curve and provide quantification by separating DNA template into ten thousands of droplets before amplification and detection, it is more tolerance to potential inhibitor [19]. dPCR method is widely used in different kinds of pathogens, including those in complex matrix [20]. Therefore, dPCR also has the potential to be used as a detection method for MPXV in clinical practice. Currently, it is very difficult to test the real clinical samples using the dPCR method, as not many native patient are infected with MPXV in China.
As the human monkeypox threatens more and more people during spreading around the world, it is crucial to ensure the comparability and validity of MPXV detection among different platforms and methods. The RM is useful for validation of the methods and kits for MPXV detection. However, due to the limit of the length of genes that can be introduced into the plasmid vector, the established Pseudovirus RM only contains the F3L gene. RMs containing other gene targets need to be further developed for evaluation of testing kits with other genes in near future.
Conclusion
MPXV leads to clinical symptoms similar to smallpox that is more fatal. MPXV can be transmitted by physical contact, respiratory spray, and polluted objects [21]. It is necessary to develop accurate higher-order reference method and RM to disease prevention and control. dPCR provides absolute quantification for nucleic acid. Based on the optimization of dPCR system, we have developed quantitative method for F3L gene of MPXV. The results indicates that our proposed dPCR method with high accuracy and good analytical sensitivity, which is proper as a high order of reference method for value assignment of the Pseudovirus RM for MPXV. The evaluation of performance of qPCR kits using our proposed RM showed that some of them cannot realize their claimed LoDs. To avoid false negative and false positive detection, it is necessary to validate the commercial kits before using in clinical diagnosis. The usage of RM can improve reliability of MPXV detection, which assists us to prevent the widespread of the infection disease caused by MPXV.
References
Petersen E, Kantele A, et al. Human monkeypox: epidemiologic and clinical characteristics, diagnosis, and prevention. Infect Dis Clin North Am. 2019;33(4):1027–43.
Fenollar F, Mediannikov O. Emerging infectious diseases in Africa in the 21st century. New Microbes New Infect. 2018;26:S10–8.
Dhawan M, Priyanka, Choudhary OP. Emergence of monkeypox: risk assessment and containment measures. Travel Med Infect Dis. 2022. 49:102392.
Rezza G. Emergence of human monkeypox in west Africa. Lancet Infect Dis. 2019;19(8):797–9.
Tian D, Zheng T. Comparison and analysis of biological agent category lists based on biosafety and biodefense. PLoS ONE. 2014;9(6): e101163.
Alakunle E, Moens U, Nchinda G, Okeke MIJV. Monkeypox virus in Nigeria: infection biology, epidemiology, and evolution. Virus. 2020;12(11):1257.
Isidro J, Borges V, Pinto M, et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus[J]. Nat Med. 2022;28(8):1569–72.
Hraib M, Jouni S, Albitar MM, Alaidi S, Alshehabi Z. The outbreak of monkeypox 2022: an overview. Ann Med Surg (Lond). 2022;79: 104069.
Shchelkunov SN, Totmenin AV, Safronov PF, Mikheev MV, Gutorov VV, Ryazankina OI, et al. Analysis of the monkeypox virus genome. Virology. 2002;297(2):172–94.
Peiró-Mestres A, Fuertes I, Camprubí-Ferrer D, et al. Frequent detection of monkeypox virus DNA in saliva, semen, and other clinical samples from 12 patients, Barcelona, Spain, May to June 2022. Euro Surveill, 2022. 27(28).
Li Y, Zhao H, Wilkins K, Hughes C, Damon IK. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J Virol Methods. 2010;169(1):223–7.
Dong L, Wang X, Wang S, et al. Interlaboratory assessment of droplet digital PCR for quantification of BRAF V600E mutation using a novel DNA reference material. Talanta. 2020;207:120293.
Niu C, Wang X, Zhang Y, et al. Interlaboratory assessment of quantification of SARS-CoV-2 RNA by reverse transcription digital PCR. Anal Bioanal Chem. 2021;413(29):7195–204.
Silva AC, Peixoto C, Lucas T, Kuppers C, Cruz PE, Alves PM, et al. Adenovirus vector production and purification. Curr Gene Ther. 2010;10(6):437–55.
IBS PD ISO GUIDE 35:2017. Reference materials — Guidance for characterization and assessment of homogeneity and stability. Switzerland: ISO; 2017.
ISO 17511:2003. In vitro diagnostic medical devices - Measurement of quantities in biological samples - Metrological traceability of values assigned to calibrators and control materials. Geneva, Switzerland: ISO; 2003.
Dong L, Zhou J, Niu C, Wang Q, Pan Y, Sheng S, et al. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. Talanta. 2021;224: 121726.
Kralik P, Ricchi M. A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front Microbiol. 2017;8:108.
Kuypers J, Jerome KR. Applications of digital PCR for clinical microbiology. J Clin Microbiol. 2017;55(6):1621–8.
Kojabad AA, Farzanehpour M, Galeh HEG, Dorostkar R, Jafarpour A, Bolandian M, et al. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J Med Virol. 2021;93(7):4182–97.
Weaver JR, Isaacs SN. Monkeypox virus and insights into its immunomodulatory proteins. Immunol Rev. 2008;225:96–113.
Funding
The research was supported by the basic research funding in key field supported by the National Institute of Metrology, People’s Republic of China (AKYZD2202 and AKYZZ2023).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, J., Guo, R., Li, H. et al. Development of highly accurate digital PCR method and reference material for monkeypox virus detection. Anal Bioanal Chem 415, 1333–1337 (2023). https://doi.org/10.1007/s00216-023-04518-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-023-04518-9