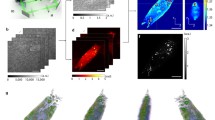
Abstract
The significance of lipid droplets in lipid metabolism, cell signaling, and regulating longevity is increasingly recognized, yet the lipid droplet’s unique properties and architecture make it difficult to size and study using conventional methods. To begin to address this issue, we demonstrate the capabilities of nanoparticle tracking analysis (NTA) for sizing of lipid droplets. NTA was found to be adequate to assess lipid droplet stability over time, indicating that lipid droplet preparations are stable for up to 24 h. NTA had the ability to compare the size distributions of lipid droplets from adult and geriatric mouse liver tissue, suggesting an age-related decrease in lipid droplet size. This is the first report on the use of NTA to size intracellular organelles.

Light scattering reveals the temporal positions of individual lipid droplets, which are recorded with a camera. The two-dimensional diffusion constant of each lipid droplet is extracted from the data set, which is then used to calculate a hydrodynamic radius using the Stokes-Einstein equation.




Similar content being viewed by others
References
Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438.
Goldberg AA, Bourque SD, Kyryakov P, Boukh-Viner T, Gregg C, Beach A, et al. A novel function of lipid droplets in regulating longevity. Biochem Soc Trans. 2009;37:1050–5.
Mcintosh AL, Storey SM, Atshaves BP. Intracellular lipid droplets contain dynamic pools of sphingomyelin: ADRP binds phospholipids with high affinity. Lipids. 2010;45:465–77.
Londos C, Brasaemle DL, Schultz CJ, Segrest JP, Kimmel AR. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin Cell Dev Biol. 1999;10:51–8.
Chanderbhan R, Noland BJ, Scallen TJ, Vahouny GV. Sterol carrier protein2. Delivery of cholesterol from adrenal lipid droplets to mitochondria for pregnanolone synthesis. J Biol Chem. 1982;257:8928–34.
Krahmer N, Farese RV, Walther TC. Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 2013;5:905–15.
Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol. 2013;10:330–44.
Day CP, James OFW. Steatohepatitis: a tale of two “Hits”? Gastroenterology. 1998;114:842–5.
Ding Y, Zhang S, Yang L, Na H, Zhang P, Zhang H, et al. Isolating lipid droplets from multiple species. Nat Protoc. 2012;8:43–51.
Nishimoto Y, Nakajima S, Tateya S, Saito M, Ogawa W, Tamori Y. Cell death-inducing DNA fragmentation factor A-like effector A and fat-specific protein 27β coordinately control lipid droplet size in brown adipocytes. J Biol Chem. 2017;292:10824–34.
Shi X, Li J, Zou X, Greggain J, Rødkær SV, Færgeman NJ, et al. Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J Lipid Res. 2013;54:2504–14.
Greenspan P, Mayer E, Fowler S. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–73.
Yang HJ, Hsu CL, Yang JY, Yang WY. Monodansylpentane as a blue-fluorescent lipid-droplet marker for multi-color live-cell imaging. PLoS One. 2012;7 https://doi.org/10.1371/journal.pone.0032693.
Mehlem A, Hagberg CE, Muhl L, Eriksson U, Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat Protoc. 2013;8:1149–55.
Klapper M, Ehmke M, Palgunow D, Böhme M, Matthäus C, Bergner G, et al. Fluorescence-based fixative and vital staining of lipid droplets in Caenorhabditis elegans reveal fat stores using microscopy and flow cytometry approaches. J Lipid Res. 2011;52:1281–93.
Deutsch MJ, Schriever SC, Roscher AA, Ensenauer R. Digital image analysis approach for lipid droplet size quantitation of oil red O-stained cultured cells. Anal Biochem. 2014;445:87–9.
Wu A, Kolanowski JL, Boumelhem BB, Yang K, Lee R, Kaur A, et al. A carborane-containing fluorophore as a stain of cellular lipid droplets. Chem Asian J. 2017;12:1704–8.
Gao M, Su H, Li S, Lin Y, Ling X, Qin A, et al. An easily accessible aggregation-induced emission probe for lipid droplet-specific imaging and movement tracking. Chem Commun. 2017;53:921–4.
Smus JP, Moura CC, McMorrow E, Tare RS, Oreffo ROC, Mahajan S. Tracking adipogenic differentiation of skeletal stem cells by label-free chemically selective imaging. Chem Sci. 2015;6:7089–96.
Cao C, Zhou D, Chen T, Streets AM, Huang Y. Label-free digital quantification of lipid droplets in single cells by stimulated raman microscopy on a microfluidic platform. Anal Chem. 2016;88:4931–9.
Kim K, Lee S, Yoon J, Heo J, Choi C, Park Y. Three-dimensional label-free imaging and quantification of lipid droplets in live hepatocytes. Sci Rep. 2016;6:1–8.
Daniele JR, Heydari K, Arriaga EA, Dillin A. Identification and characterization of mitochondrial subtypes in caenorhabditis elegans via analysis of individual mitochondria by flow cytometry. Anal Chem. 2016;88:6309–16.
Taylor TH, Frost NW, Bowser MT, Arriaga EA. Analysis of individual mitochondria via fluorescent immunolabeling with anti-TOM22 antibodies. Anal Bioanal Chem. 2014;406:1683–91.
Degtyarev M, Reichelt M, Lin K. Novel quantitative autophagy analysis by organelle flow cytometry after cell sonication. PLoS One. 2014;9 https://doi.org/10.1371/journal.pone.0087707.
Malloy A, Carr B. Nanoparticle tracking analysis - the halo system. Part Syst Charact. 2006;23:197–204.
Soo CY, Song Y, Zheng Y, Campbell EC, Riches AC, Gunn-Moore F, et al. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136:192–7.
Oosthuyzen W, Sime NEL, Ivy JR, Turtle EJ, Street JM, Pound J, et al. Quantification of human urinary exosomes by nanoparticle tracking analysis. J Physiol. 2013;23:5833–42.
Tatischeff I, Larquet E, Falcon-Perez JM, Turpin PY, Kruglik SG (2012) Fast characterization of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. J Extracell Vesicles 1: doi: https://doi.org/10.3402/jev.v1i0.19179
Gardiner C, Ferreira YJ, Dragovic RA, Redman CWG, Sargent IL. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J Extracell Vesicles. 2013;2 https://doi.org/10.3402/jev.v2i0.19671.
Kramberger P, Ciringer M, Štrancar A, Peterka M. Evaluation of nanoparticle tracking analysis for total virus particle determination. Virol J. 2012;9:265.
James AE, Driskell JD. Monitoring gold nanoparticle conjugation and analysis of biomolecular binding with nanoparticle tracking analysis (NTA) and dynamic light scattering (DLS). Analyst. 2013;138:1212.
Atshaves BP, Storey SM, McIntosh AL, Petrescu AD, Lyuksyutova OI, Greenberg AS, et al. Sterol carrier protein-2 expression modulates protein and lipid composition of lipid droplets. J Biol Chem. 2001;276:25324–35.
Storey SM, AL MI, Senthivinayagam S, Moon KC, Atshaves BP. The phospholipid monolayer associated with perilipin-enriched lipid droplets is a highly organized rigid membrane structure. AJP Endocrinol Metab. 2011;301:E991–E1003.
Chandrasekhar S. Brownian motion, dynamical friction, and stellar dynamics. Rev Mod Phys. 1949;21:383–8.
Einstein A. On the motion of small particles suspended in a stationary liquid, as required by the molecular kinetic theory of heat. Ann Phys. 1905;322:549–60.
Marx E, Mulholland GW. Size and refractive index determination of single polystyrene spheres. J Res Natl Bur Stand. 1983;88(5):321.
Haseda K, Kanematsu K, Noguchi K, Saito H, Umeda N, Ohta Y. Significant correlation between refractive index and activity of mitochondria: single mitochondrion study. Biomed Opt Express. 2015;6:859.
Gouw TH, Vlugter JC. Physical properties of triglycerides. !. Density and refractive index. Eur J Lipid Sci Technol. 1966;68:544–69.
Suzuki M, Shinohara Y, Ohsaki Y, Fujimoto T (2011) Lipid droplets: Size matters. J Electron Microsc 60: https://doi.org/10.1093/jmicro/dfr016
Najt CP, Senthivinayagam S, Aljazi MB, Fader KA, Olenic SD, Brock JRL, et al. Liver-specific loss of perilipin 2 alleviates diet-induced hepatic steatosis, inflammation, and fibrosis. Am J Physiol Gastrointest Liver Physiol. 2016;310:G726–38.
McManaman JL, Bales ES, Orlicky DJ, Jackman M, MacLean PS, Cain S, et al. Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J Lipid Res. 2013;54:1346–59.
Zhang S, Wang Y, Cui L, Deng Y, Xu S, Yu J, et al. Morphologically and functionally distinct lipid droplet subpopulations. Sci Rep. 2016;6:29539.
Cohen B-C, Shamay A, Argov-Argaman N. Regulation of lipid droplet size in mammary epithelial cells by remodeling of membrane lipid composition-a potential mechanism. PLoS One. 2015;10:e0121645.
Bonda-Ostaszewska E, Włostowski T, Krasowska A, Kozłowski P. Seasonal and photoperiodic effects on lipid droplet size and lipid peroxidation in the brown adipose tissue of bank voles (Myodes glareolus). Acta Theriol (Warsz). 2012;57:289–94.
Slawik M, Vidal-Puig AJ. Lipotoxicity, overnutrition, and energy metabolism in aging. Aging Res Rev. 2006;5:144–64.
Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, et al. Mitochondrial dysfunction in the elderly: Possible role in insulin resistance. Science. 2003;300(80):1140–2.
Daniele JR, Esping DJ, Garcia G, Parsons LS, Arriaga EA, Dillin A. High-throughput characterization of region-specific mitochondrial function and morphology. Sci Rep. 2017;7:6749.
Funding
This work was supported by NIH AG020866. KAM acknowledges support through a University of Minnesota Doctoral Dissertation Fellowship and National Institutes of Health (NIH) AG029796. CPN acknowledges support from NIH DK007203. NML acknowledges support from NIH GM008700. DGM acknowledges support from NIH DK114401 and NIH AG055452. A portion of this work was carried out in the Minnesota Nano Center, which receives partial support from the National Science Foundation (NSF) through the NNCI program. A portion of this work was carried out in the Characterization Facility, University of Minnesota, which receives partial support from NSF through the MRSEC program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All mice were housed in a designated clean facility and treated in accordance with protocols approved by the University of Minnesota Institutional Animal Care and Use Committee.
Conflict of Interest
The authors have no potential conflicts of interest to disclose.
Rights and permissions
About this article
Cite this article
Muratore, K.A., Najt, C.P., Livezey, N.M. et al. Sizing lipid droplets from adult and geriatric mouse liver tissue via nanoparticle tracking analysis. Anal Bioanal Chem 410, 3629–3638 (2018). https://doi.org/10.1007/s00216-018-1016-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1016-8