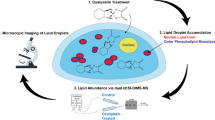
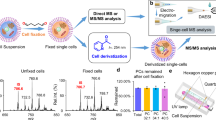
Abstract
The importance of the polo-like kinase 1 (PLK1) gene is increasing substantially both as a biomarker and as a target for highly specific cancer therapy. This is due to its involvement in multiple points of cell progression and carcinogenesis. PLK1 inhibitors’ efficacy in treating human cancers has been limited due to the lack of a specific targeting strategy. Here, we describe a method of targeted downregulation of PLK1 in cancer cells and the concomitant rapid detection of surface lipidomic perturbations using desorption electrospray ionization mass spectrometry (DESI MS). The efficient delivery of siRNA targeting PLK1 gene selectively to the cancer cells is achieved by targeting overexpressed cell surface epithelial cell adhesion molecule (EpCAM) by the EpDT3 aptamer. The chimeric aptamer (EpDT3-siPLK1) showed the knockdown of PLK1 gene expression and PLK1 protein levels by quantitative PCR and western blotting, respectively. The abundant surface lipids, phosphatidylcholines (PCs), such as PC(32:1) (m/z 754.6), PC(34:1) (m/z 782.6), and PC(36:2) (m/z 808.6), were highly expressed in MCF-7 and WERI-RB1 cancer cells compared to normal MIO-M1 cells and they were observed using DESI MS. These overexpressed cell surface lipids in the cancer cells were downregulated upon the treatment of EpDT3-siPLK1 chimera indicating a novel role of PLK1 to regulate surface lipid expression in addition to the efficient selective cancer targeting ability. Our results indicate that DESI MS has a potential ability to rapidly monitor aptamer-mediated cancer therapy and accelerate the drug discovery process.

Binding of aptamer chimera to the cells and changes in lipid profile





Similar content being viewed by others
References
Beljebbar A, Dukic S, Amharref N, Bellefqih S, Manfait M. Monitoring of biochemical changes through the C6 gliomas progression and invasion by Fourier transform infrared (FTIR) imaging. Anal Chem. 2009;81:9247–56.
Shimma S, Sugiura Y, Hayasaka T, Hoshikawa Y, Noda T, Setou M. MALDI-based imaging mass spectrometry revealed abnormal distribution of phospholipids in colon cancer liver metastasis. J Chromatogr B. 2007;855:98–103.
Ackerstaff E, Glunde K, Bhujwalla ZM. Choline phospholipid metabolism: a target in cancer cells? J Cell Biochem. 2003;90:525–33.
Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–6.
Gillies RJ, Morse DL. In vivo magnetic resonance spectroscopy in cancer. Annu Rev Biomed Eng. 2005;7:287–326.
Glunde K, Artemov D, Penet M-F, Jacobs MA, Bhujwalla ZM. Magnetic resonance spectroscopy in metabolic and molecular imaging and diagnosis of cancer. Chem Rev. 2010;110:3043–59.
Monge ME, Harris GA, Dwivedi P, Fernandez FM. Mass spectrometry: recent advances in direct open air surface sampling/ionization. Chem Rev. 2013;113:2269–308.
Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–3.
Pasilis SP, Kertesz V, Van Berkel GJ, Schulz M, Schorcht S. HPTLC/DESI-MS imaging of tryptic protein digests separated in two dimensions. J Mass Spectrom. 2008;43:1627–35.
Chipuk JE, Gelb MH, Brodbelt JS. Rapid and selective screening for sulfhydryl analytes in plasma and urine using surface-enhanced transmission mode desorption electrospray ionization mass spectrometry. Anal Chem. 2010;82:4130–9.
Wiseman JM, Ifa DR, Venter A, Cooks RG. Ambient molecular imaging by desorption electrospray ionization mass spectrometry. Nat Protoc. 2008;3:517–24.
Vismeh R, Waldon DJ, Teffera Y, Zhao Z. Localization and quantification of drugs in animal tissues by use of desorption electrospray ionization mass spectrometry imaging. Anal Chem. 2012;84:5439–45.
Eberlin LS, Norton I, Orringer D, Dunn IF, Liu X, Ide JL, et al. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc Natl Acad Sci U S A. 2013;110:1611–6.
Eberlin LS, Tibshirani RJ, Zhang J, Longacre TA, Berry GJ, Bingham DB, et al. Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc Natl Acad Sci U S A. 2014;111:2436–41.
Srimany A, Ifa DR, Naik HR, Bhat V, Cooks RG, Pradeep T. Direct analysis of camptothecin from Nothapodytes nimmoniana by desorption electrospray ionization mass spectrometry (DESI-MS). Analyst. 2011;136:3066–8.
Ifa DR, Srimany A, Eberlin LS, Naik HR, Bhat V, Cooks RG, et al. Tissue imprint imaging by desorption electrospray ionization mass spectrometry. Anal Methods. 2011;3:1910–2.
Mohana Kumara P, Srimany A, Ravikanth G, Uma Shaanker R, Pradeep T. Ambient ionization mass spectrometry imaging of rohitukine, a chromone anti-cancer alkaloid, during seed development in Dysoxylum binectariferum Hook.f (Meliaceae). Phytochemistry. 2015;116:104–10.
Hemalatha RG, Pradeep T. Understanding the molecular signatures in leaves and flowers by desorption electrospray ionization mass spectrometry (DESI MS) imaging. J Agric Food Chem. 2013;61:7477–87.
Glover DM, Hagan IM, Tavares ÁAM. Polo-like kinases: a team that plays throughout mitosis. Gene Dev. 1998;12:3777–87.
Glover DM, Ohkura H, Tavares A. Polo kinase: the choreographer of the mitotic stage? J Cell Biol. 1996;135:1681–4.
Nigg EA, Blangy A, Lane HA. Dynamic changes in nuclear architecture during mitosis: on the role of protein phosphorylation in spindle assembly and chromosome segregation. Exp Cell Res. 1996;229:174–80.
Kumagai A, Dunphy WG. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science. 1996;273:1377–80.
Nigg EA. Polo-like kinases: positive regulators of cell division from start to finish. Curr Opin Cell Biol. 1998;10:776–83.
Cunningham JT, Ruggero D. New connections between old pathways: PDK1 signaling promotes cellular transformation through PLK1-dependent MYC stabilization. Cancer Discov. 2013;3:1099–102.
Wang G, Chen Q, Zhang X, Zhang B, Zhuo X, Liu J, et al. PCM1 recruits Plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J Cell Sci. 2013;126:1355–65.
Cholewa BD, Liu X, Ahmad N. The role of polo-like kinase 1 in carcinogenesis: cause or consequence? Cancer Res. 2013;73:6848–55.
Bhola NE, Jansen VM, Bafna S, Giltnane JM, Balko JM, Estrada MV, et al. Kinome-wide functional screen identifies role of PLK1 in hormone-independent, ER-positive breast cancer. Cancer Res. 2015;75:405–14.
Zhang Z, Hou X, Shao C, Li J, Cheng J-X, Kuang S, et al. Plk1 inhibition enhances the efficacy of androgen signaling blockade in castration-resistant prostate cancer. Cancer Res. 2014;74:6635–47.
Denholt CL, Hansen PR, Pedersen N, Poulsen HS, Gillings N, Kjær A. Identification of novel peptide ligands for the cancer-specific receptor mutation EFGRvIII using a mixture-based synthetic combinatorial library. Biopolymers. 2009;91:201–6.
Kim M, Shin D-S, Kim J, Lee Y-S. Substrate screening of protein kinases: detection methods and combinatorial peptide libraries. Biopolymers. 2010;94:753–62.
Li J, Tan S, Chen X, Zhang CY, Zhang Y. Peptide aptamers with biological and therapeutic applications. Curr Med Chem. 2011;18:4215–22.
Pirogova E, Istivan T, Gan E, Cosic I. Advances in methods for therapeutic peptide discovery, design and development. Curr Pharm Biotechnol. 2011;12:1117–27.
Aina OH, Liu R, Sutcliffe JL, Marik J, Pan C-X, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol Pharmaceutics. 2007;4:631–51.
Zhu Z, Song Y, Li C, Zou Y, Zhu L, An Y, et al. Monoclonal surface display SELEX for simple, rapid, efficient, and cost-effective aptamer enrichment and identification. Anal Chem. 2014;86:5881–8.
Subramanian N, Raghunathan V, Kanwar JR, Kanwar RK, Elchuri SV, Khetan V, et al. Target-specific delivery of doxorubicin to retinoblastoma using epithelial cell adhesion molecule aptamer. Mol Vis. 2012;18:2783–95.
Gilboa-Geffen A, Hamar P, Le MTN, Wheeler LA, Trifonova R, Petrocca F, et al. Gene knockdown by EpCAM aptamer-siRNA chimeras suppresses epithelial breast cancers and their tumor-initiating cells. Mol Cancer Ther. 2015;14:2279–91.
Srimany A, Jayashree B, Krishnakumar S, Elchuri S, Pradeep T. Identification of effective substrates for the direct analysis of lipids from cell lines using desorption electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2015;29:349–56.
Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27:839–49.
Subramanian N, Kanwar JR, Athalya PK, Janakiraman N, Khetan V, Kanwar RK, et al. EpCAM aptamer mediated cancer cell specific delivery of EpCAM siRNA using polymeric nanocomplex. J Biomed Sci. 2015;22:4.
Liu HY, Gao X. A universal protein tag for delivery of SiRNA-aptamer chimeras. Sci Rep. 2013;3:3129.
Zhou J, Tiemann K, Chomchan P, Alluin J, Swiderski P, Burnett J, et al. Dual functional BAFF receptor aptamers inhibit ligand-induced proliferation and deliver siRNAs to NHL cells. Nucleic Acids Res. 2013;41:4266–83.
Eberlin LS, Gabay M, Fan AC, Gouw AM, Tibshirani RJ, Felsher DW, et al. Alteration of the lipid profile in lymphomas induced by MYC overexpression. Proc Natl Acad Sci U S A. 2014;111:10450–5.
Yang L, Cui X, Zhang N, Li M, Bai Y, Han X, et al. Comprehensive lipid profiling of plasma in patients with benign breast tumor and breast cancer reveals novel biomarkers. Anal Bioanal Chem. 2015;407:5065–77.
Liu R, Peng Y, Li X, Wang Y, Pan E, Guo W, et al. Identification of plasma metabolomic profiling for diagnosis of esophageal squamous-cell carcinoma using an UPLC/TOF/MS platform. Int J Mol Sci. 2013;14:8899–911.
Huang C, Freter C. Lipid metabolism, apoptosis and cancer therapy. Int J Mol Sci. 2015;16:924–49.
Acknowledgments
This work was supported by the Department of Biotechnology, Government of India (Grant no. NO.BT/PR3580/PID/6/625/2011) and the Department of Science and Technology, Government of India. A.S. thanks the Council of Scientific and Industrial Research, Government of India and Indian Institute of Technology Madras for fellowships. The authors also thank Dr. Nithya Subramanian, Ms. Srilatha Jasty, and Ms. Lakshmi for their help and suggestions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Balasubramanyam Jayashree and Amitava Srimany contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 0.99 mb)
Rights and permissions
About this article
Cite this article
Jayashree, B., Srimany, A., Jayaraman, S. et al. Monitoring of changes in lipid profiles during PLK1 knockdown in cancer cells using DESI MS. Anal Bioanal Chem 408, 5623–5632 (2016). https://doi.org/10.1007/s00216-016-9665-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9665-y