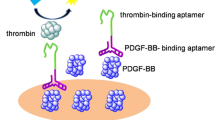
Abstract
We describe a sensitive aptamer-based sandwich assay for protein detection on microplate by using rolling circle amplification (RCA) coupled with thrombin catalysis. This assay takes advantage of RCA generating long DNA oligonucleotides with repeat thrombin-binding aptamer sequence, specific aptamer affinity binding to achieve multiple thrombin labeling, and enzyme activity of thrombin for signal generation. Protein target is specifically captured by antibody-coated microplate. Then, an oligonucleotide containing an aptamer for protein and a primer sequence is added to form a typical sandwich structure. Following a template encoded with complementary sequence of aptamer for thrombin, RCA reaction extends the primer sequence into a long oligonucleotide. Many thrombin molecules bind with the RCA product. Thrombin catalyzes the conversion of its chromogenic or fluorogenic peptide substrates into detectable products for final quantification of protein targets. We applied this strategy to the detection of a model protein target, platelet-derived growth factor-BB (PDGF-BB). Due to double signal amplifications from RCA and thrombin catalysis, this assay enabled the detection of PDGF-BB as low as 3.1 pM when a fluorogenic peptide substrate was used. This assay provides a new way for signal generation in RCA-involved assay through direct thrombin labeling, circumventing time-consuming preparation of enzyme-conjugate and affinity probes. This method has promise for a variety of analytical applications.






Similar content being viewed by others
References
Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–22.
Tuerk C, Gold L. Systemic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–10.
Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287(5454):820–5.
Li F, Zhang H, Wang Z, Newbigging AM, Reid MS, Li XF, et al. Aptamers facilitating amplified detection of biomolecules. Anal Chem. 2015;87(1):274–92.
Liu J, Cao Z, Lu Y. Functional nucleic acids sensors. Chem Rev. 2009;109(5):1948–98.
Juskowiak B. Nucleic acid-based fluorescent probes and their analytical potential. Anal Bioanal Chem. 2011;399(9):3157–76.
Cho EJ, Lee JW, Ellington AD. Applications of aptamers as sensors. Annu Rev Anal Chem. 2009;2:241–64.
Zhou J, Battig MR, Wang Y. Aptamer-based molecular recognition for biosensor development. Anal Bioanal Chem. 2010;398(6):2471–80.
Deng N, Liang Z, Liang Y, Sui Z, Zhang L, Wu Q, et al. Aptamer modified organic–inorganic hybrid silica monolithic capillary columns for highly selective recognition of thrombin. Anal Chem. 2012;84(23):10186–90.
Zhang H, Li F, Dever B, Li XF, Le XC. DNA-mediated homogeneous binding assays for nucleic acids and proteins. Chem Rev. 2013;113(4):2812–41.
Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115(22):12491–545.
Ali MM, Li F, Zhang ZQ, Zhang KX, Kang DK, Ankrum JA, et al. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem Soc Rev. 2014;43(10):3324–41.
Schweitzer B, Wiltshire S, Lambert J, O’Malley S, Kukanskis K, Zhu Z, et al. Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc Natl Acad Sci U S A. 2000;97(18):10113–9.
Zhou L, Ou LJ, Chu X, Shen GL, Yu RQ. Aptamer-based rolling circle amplification: a platform for electrochemical detection of protein. Anal Chem. 2007;79(19):7942–500.
Bi S, Li L, Zhang SS. Triggered polycatenated DNA scaffolds for DNA sensors and aptasensors by a combination of rolling circle amplification and DNAzyme amplification. Anal Chem. 2010;82(22):9447–54.
Tang LH, Liu Y, Ali MM, Kang D, Zhao WA, Li JH. Colorimetric and ultrasensitive bioassay based on a dual-amplification system using aptamer and DNAzyme. Anal Chem. 2012;84(11):4711–7.
Lee J, Icoz K, Roberts A, Ellington AD, Savran CA. Diffractometric detection of proteins using microbead-based rolling circle amplification. Anal Chem. 2010;82(1):197–202.
Yang LT, Fung CW, Cho EJ, Ellington AD. Real-time rolling circle amplification for protein detection. Anal Chem. 2007;79(9):3320–9.
Wang QP, Zheng HY, Gao XY, Lin ZY, Chen GN. A label-free ultrasensitive electrochemical aptameric recognition system for protein assay based on hyperbranched rolling circle amplification. Chem Commun. 2013;49(97):11418–20.
Cao ZJ, Peng QW, Qiu X, Liu CY, Lu JZ. Highly sensitive chemiluminescence technology for protein detection using aptamer-based rolling circle amplification platform. J Pharm Anal. 2011;1(3):159–65.
Wang L, Tram K, Ali MM, Salena BJ, Li J, Li Y. Arrest of rolling circle amplification by protein-binding DNA aptamers. Chem Eur J. 2014;20(9):2420–4.
Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355(6360):564–6.
Tasset DM, Kubik MF, Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J Mol Biol. 1997;272(5):688–98.
Deng B, Lin Y, Wang C, Li F, Wang Z, Zhang H, et al. Aptamer binding assays for proteins: the thrombin example—a review. Anal Chim Acta. 2014;837:1–15.
Wu Q, Tsiang M, Sadler JE. Localization of the single-stranded DNA binding site in the thrombin anion-binding exosite. J Bio Chem. 1992;267(34):24408–12.
Zhou G, Huang X, Qu Y. The binding effect of aptamers on thrombin. Biochem Eng J. 2010;52(2–3):117–22.
Mir M, Vreeke M, Katakis I. Different strategies to develop an electrochemical thrombin aptasensor. Electrochem Commun. 2006;8(3):505–11.
Muller J, Becher T, Braunstein J, Berdel P, Gravius S, Rohrbach F, et al. Profiling of active thrombin in human blood by supramolecular complexes. Angew Chem Int Ed. 2011;50(27):6075–8.
Zhao Q, Li XF, Le XC. Aptamer capturing of enzymes on magnetic beads to enhance assay specificity and sensitivity. Anal Chem. 2011;83(24):9234–6.
Zhao Q, Wang XF. An aptamer-capture based chromogenic assay for thrombin. Biosens Bioelectron. 2012;34(1):232–7.
Westermark B, Heldin CH. Platelet-derived growth factor. Structure, function and implications in normal and malignant cell growth. Acta Oncol. 1993;32(2):101–5.
Heldin CH. Structural and functional studies on platelet-derived growth factor. The EMBO J. 1992;11(12):4251–9.
Green LS, Jellinek D, Jenison R, Ostman A, Heldin CH, Janjic N. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry. 1996;35(45):14413–24.
Jin X, Zhao JJ, Zhang LL, Huang Y, Zhao SL. An enhanced fluorescence polarization strategy based on multiple protein–DNA–protein structures for sensitive detection of PDGF-BB. RSC Adv. 2014;4(13):6850–3.
Li H, Zhu Y, Dong SY, Qiang WB, Sun L, Xu DK. Fast functionalization of silver decahedral nanoparticles with aptamers for colorimetric detection of human platelet-derived growth factor-BB. Anal Chim Acta. 2014;829:48–53.
Wang P, Song YH, Zhao YJ, Fan AP. Hydroxylamine amplified gold nanoparticle-based aptameric system for highly selective and sensitive detection of platelet-derived growth factor. Talanta. 2013;103:392–7.
Chang CC, Wei SC, Wu TH, Lee CH, Lin CW. Apatamer-based colorimetric detection of platelet-derived growth factor using unmodified gold nanoparticles. Biosens Bioelectron. 2013;42:119–23.
Fang XH, Li JJ, Tan WH. Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal Chem. 2000;72(14):3280–5.
Zhang H, Li XF, Le XC. Differentiation and detection of PDGF isomers and their receptors by tunable aptamer capillary electrophoresis. Anal Chem. 2009;81(18):7795–800.
Acknowledgments
We thank the support from National Natural Science Foundation of China (Grant No. 21222503) and Outstanding Youth Talents Program of Shanxi Province.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 205 kb)
Rights and permissions
About this article
Cite this article
Guo, L., Hao, L. & Zhao, Q. An aptamer assay using rolling circle amplification coupled with thrombin catalysis for protein detection. Anal Bioanal Chem 408, 4715–4722 (2016). https://doi.org/10.1007/s00216-016-9558-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9558-0