Abstract
Quantitative methods for detection of biological molecules are needed more than ever before in the emerging age of “omics” and “big data.” Here, we provide an integrated approach for systematic analysis of the “lipidome” in tissue. To test our approach in a biological context, we utilized brain tissue selectively deficient for the transcription factor Specificity Protein 2 (Sp2). Conditional deletion of Sp2 in the mouse cerebral cortex results in developmental deficiencies including disruption of lipid metabolism. Silver (Ag) cationization was implemented for infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) to enhance the ion abundances for olefinic lipids, as these have been linked to regulation by Sp2. Combining Ag-doped and conventional IR-MALDESI imaging, this approach was extended to IR-MALDESI imaging of embryonic mouse brains. Further, our imaging technique was combined with bottom-up shotgun proteomic LC-MS/MS analysis and western blot for comparing Sp2 conditional knockout (Sp2-cKO) and wild-type (WT) cortices of tissue sections. This provided an integrated omics dataset which revealed many specific changes to fundamental cellular processes and biosynthetic pathways. In particular, step-specific altered abundances of nucleotides, lipids, and associated proteins were observed in the cerebral cortices of Sp2-cKO embryos.

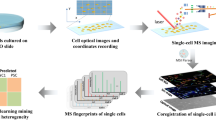
TransOmic Analysis of Dorsolateral cortices of Sp2 conditional Knockout mouse embryos. Target tissue was extracted by laser microdissection and analyzed by LC-MS/MS label- free quantitative proteomics. In parallel, lipid imaging of these tissues was performed by conventional IR-MALDESI and Agdoped IR-MALDESI imaging. These data were then integrated to obtain insight into lipid pathways altered by presence or absence of transcription factor Specificity Protein 2










Similar content being viewed by others
References
Volkel S, Stielow B, Finkernagel F, Stiewe T, Nist A, Suske G. Zinc finger independent genome-wide binding of Sp2 potentiates recruitment of histone-fold protein Nf-y distinguishing it from Sp1 and Sp3. PLoS Genet. 2015;3, e1005102.
Terrados G, Finkernagel F, Stielow B, Sadic D, Neubert J, Herdt O, et al. Genome-wide localization and expression profiling establish Sp2 as a sequence-specific transcription factor regulating vitally important genes. Nucleic Acids Res. 2012;16:7844–57.
Liang H, Xiao G, Yin H, Hippenmeyer S, Horowitz JM, Ghashghaei HT. Neural development is dependent on the function of specificity protein 2 in cell cycle progression. Development. 2013;3:552–61.
Robichaud G, Barry JA, Garrard KP, Muddiman DC. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J Am Soc Mass Spectrom. 2013;1:92–100.
Robichaud G, Barry JA, Muddiman DC. IR-MALDESI mass spectrometry imaging of biological tissue sections using ice as a matrix. J Am Soc Mass Spectrom. 2014;3:319–28.
Barry JA, Muddiman DC. Global optimization of the infrared matrix-assisted laser desorption electrospray ionization (IR MALDESI) source for mass spectrometry using statistical design of experiments. Rapid Commun Mass Spectrom. 2011;23:3527–36.
Houjou T, Yamatani K, Imagawa M, Shimizu T, Taguchi R. A shotgun tandem mass spectrometric analysis of phospholipids with normal-phase and/or reverse-phase liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2005;5:654–66.
Shoji N, Nakagawa K, Asai A, Fujita I, Hashiura A, Nakajima Y, et al. LC-MS/MS analysis of carboxymethylated and carboxyethylated phosphatidylethanolamines in human erythrocytes and blood plasma. J Lipid Res. 2010;8:2445–53.
Myers DS, Ivanova PT, Milne SB, Brown HA. Quantitative analysis of glycerophospholipids by LC-MS: acquisition, data handling, and interpretation. Biochim Biophys Acta Mol Cell Biol Lipids. 2011;11:748–57.
Ecker J. Profiling eicosanoids and phospholipids using LC-MS/MS: principles and recent applications. J Sep Sci. 2012;10–11:1227–35.
Ogiso H, Suzuki T, Taguchi R. Development of a reverse-phase liquid chromatography electrospray ionization mass spectrometry method for lipidomics, improving detection of phosphatidic acid and phosphatidylserine. Anal Biochem. 2008;1:124–31.
Roberg-Larsen H, Vesterdal C, Wilson SR, Lundanes E. Underivatized oxysterols and nanoLC-ESI-MS: a mismatch. Steroids. 2015;99:125–30.
Fhaner CJ, Liu SC, Ji H, Simpson RJ, Reid GE. Comprehensive lipidome profiling of isogenic primary and metastatic colon adenocarcinoma cell lines. Anal Chem. 2012;21:8917–26.
Meier F, Garrard KP, Muddiman DC. Silver dopants for targeted and untargeted direct analysis of unsaturated lipids via infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Rapid Commun Mass Spectrom. 2014;22:2461–70.
Gokce E, Franck WL, Oh Y, Dean RA, Muddiman DC. In-depth analysis of the Magnaporthe oryzae conidial proteome. J Proteome Res. 2012;12:5827–35.
Loziuk PL, Parker J, Li W, Lin CY, Wang JP, Li Q, et al. Elucidation of xylem-specific transcription factors and absolute quantification of enzymes regulating cellulose biosynthesis in Populus trichocarpa. J Proteome Res. 2015.
Scheltema RA, Hauschild JP, Lange O, Hornburg D, Denisov E, Damoc E, et al. The Q exactive HF, a benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol Cell Proteomics. 2014;12:3698–708.
Kelstrup CD, Jersie-Christensen RR, Batth TS, Arrey TN, Kuehn A, Kellmann M, et al. Rapid and deep proteomes by faster sequencing on a benchtop quadrupole ultra-high-field Orbitrap mass spectrometer. J Proteome Res. 2014;12:6187–95.
Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;4:1017–31.
Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;12:2010–21.
Jones EA, Deininger SO, Hogendoorn PC, Deelder AM, McDonnell LA. Imaging mass spectrometry statistical analysis. J Proteome. 2012;16:4962–89.
Fonville JM, Carter C, Cloarec O, Nicholson JK, Lindon JC, Bunch J, et al. Robust data processing and normalization strategy for MALDI mass spectrometric imaging. Anal Chem. 2012;3:1310–9.
Bray JH, Scott E. Multivariate analysis of variance. Newbury park, CA: Sage; 1985.
Krzywinski M, Altman N. Significance, P values and t-tests. Nat Methods. 2013;11:1041–2.
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;1–2:279–84.
Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;6:747–51.
Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;9:R183.
Dietschy JM, Turley SD. Thematic review series: brain lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J Lipid Res. 2004;8:1375–97.
Bjorkhem I, Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler Thromb Vasc Biol. 2004;5:806–15.
van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;2:112–24.
Orth M, Bellosta S. Cholesterol: its regulation and role in central nervous system disorders. Cholesterol. 2012;292598.
Herz J, Farese Jr RV. The LDL receptor gene family, apolipoprotein B and cholesterol in embryonic development. J Nutr. 1999;2S(Suppl):473S–5S.
Saher G, Brugger B, Lappe-Siefke C, Mobius W, Tozawa R, Wehr MC, et al. High cholesterol level is essential for myelin membrane growth. Nat Neurosci. 2005;4:468–75.
Lanekoff I, Burnum-Johnson K, Thomas M, Short J, Carson JP, Cha J, et al. High-speed tandem mass spectrometric in situ imaging by nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2013;20:9596–603.
Landgraf RR, Prieto Conaway MC, Garrett TJ, Stacpoole PW, Yost RA. Imaging of lipids in spinal cord using intermediate pressure matrix-assisted laser desorption-linear ion trap/Orbitrap MS. Anal Chem. 2009;20:8488–95.
Perdian DC, Lee YJ. Imaging MS methodology for more chemical information in less data acquisition time utilizing a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2010;22:9393–400.
Nemes P, Woods AS, Vertes A. Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2010;3:982–8.
Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem Rev. 2011;10:6491–512.
Gode D, Volmer DA. Lipid imaging by mass spectrometry—a review. Analyst. 2013;5:1289–315.
Nazari M, Muddiman DC. Polarity switching mass spectrometry imaging of healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Analyst. 2015.
Laboratories, K. Steroid biosynthesis, KEGG pathway 00100, version 11/29/13. Available: http://www.genome.jp/kegg-bin/show_pathway?map00100
Matusch A, Fenn LS, Depboylu C, Klietz M, Strohmer S, McLean JA, et al. Combined elemental and biomolecular mass spectrometry imaging for probing the inventory of tissue at a micrometer scale. Anal Chem. 2012;7:3170–8.
Berridge MJ. Inositol trisphosphate and diacylglycerol as second messengers. Biochem J. 1984;2:345–60.
Berridge MJ. Inositol trisphosphate and diacylglycerol: two interacting second messengers. Annu Rev Biochem. 1987;56:159–93.
Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 1995;7:484–96.
Adachi N, Oyasu M, Taniguchi T, Yamaguchi Y, Takenaka R, Shirai Y, et al. Immunocytochemical localization of a neuron-specific diacylglycerol kinase beta and gamma in the developing rat brain. Brain Res Mol Brain Res. 2005;2:288–99.
Quan G, Xie C, Dietschy JM, Turley SD. Ontogenesis and regulation of cholesterol metabolism in the central nervous system of the mouse. Brain Res Dev Brain Res. 2003;1–2:87–98.
Hankin JA, Farias SE, Barkley RM, Heidenreich K, Frey LC, Hamazaki K, et al. MALDI mass spectrometric imaging of lipids in rat brain injury models. J Am Soc Mass Spectrom. 2011;6:1014–21.
Sugiura Y, Konishi Y, Zaima N, Kajihara S, Nakanishi H, Taguchi R, et al. Visualization of the cell-selective distribution of PUFA-containing phosphatidylcholines in mouse brain by imaging mass spectrometry. J Lipid Res. 2009;9:1776–88.
Li F, Qin XZ, Chen HQ, Qiu L, Guo YM, Liu H, et al. Lipid profiling for early diagnosis and progression of colorectal cancer using direct-infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 2013;1:24–34.
Tucci S, Flogel U, Spiekerkoetter U. Sexual dimorphism of lipid metabolism in very long-chain acyl-CoA dehydrogenase deficient (VLCAD(−/−)) mice in response to medium-chain triglycerides (MCT). Biochim BiophysActa (BBA) - Mol Basis Dis. 2015;7:1442–50.
Jenkins B, West JA, Koulman A. A review of odd-chain fatty acid metabolism and the role of pentadecanoic acid (C15:0) and heptadecanoic acid (C17:0) in health and disease. Molecules. 2015;2:2425–44.
Alexandrov T, Becker M, Deininger SO, Ernst G, Wehder L, Grasmair M, et al. Spatial segmentation of imaging mass spectrometry data with edge-preserving image denoising and clustering. J Proteome Res. 2010;12:6535–46.
Alexandrov T, Kobarg JH. Efficient spatial segmentation of large imaging mass spectrometry datasets with spatially aware clustering. Bioinformatics. 2011;13:i230–8.
Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–32. Database issue.
Deacon TW, Pakzaban P, Isacson O. The lateral ganglionic eminence is the origin of cells committed to striatal phenotypes: neural transplantation and developmental evidence. Brain Res. 1994;1–2:211–9.
de Carlos JA, Lopez-Mascaraque L, Valverde F. Dynamics of cell migration from the lateral ganglionic eminence in the rat. J Neurosci. 1996;19:6146–56.
Wichterle H, Garcia-Verdugo JM, Herrera DG, Alvarez-Buylla A. Young neurons from medial ganglionic eminence disperse in adult and embryonic brain. Nat Neurosci. 1999;5:461–6.
van Meer G. Cellular lipidomics. EMBO J. 2005;18:3159–65.
Saito K, Dubreuil V, Arai Y, Wilsch-Brauninger M, Schwudke D, Saher G, et al. Ablation of cholesterol biosynthesis in neural stem cells increases their VEGF expression and angiogenesis but causes neuron apoptosis. Proc Natl Acad Sci U S A. 2009;20:8350–5.
Andreyev AY, Fahy E, Guan Z, Kelly S, Li X, McDonald JG, et al. Subcellular organelle lipidomics in TLR-4-activated macrophages. J Lipid Res. 2010;9:2785–97.
Tanyi JL, Hasegawa Y, Lapushin R, Morris AJ, Wolf JK, Berchuck A, et al. Role of decreased levels of lipid phosphate phosphatase-1 in accumulation of lysophosphatidic acid in ovarian cancer. Clin Cancer Res. 2003;10(Pt 1):3534–45.
Murray NR, Fields AP. Phosphatidylglycerol is a physiologic activator of nuclear protein kinase C. J Biol Chem. 1998;19:11514–20.
Jokela H, Rantakari P, Lamminen T, Strauss L, Ola R, Mutka AL, et al. Hydroxysteroid (17beta) dehydrogenase 7 activity is essential for fetal de novo cholesterol synthesis and for neuroectodermal survival and cardiovascular differentiation in early mouse embryos. Endocrinology. 2010;4:1884–92.
Rantakari P, Lagerbohm H, Kaimainen M, Suomela JP, Strauss L, Sainio K, et al. Hydroxysteroid (17{beta}) dehydrogenase 12 is essential for mouse organogenesis and embryonic survival. Endocrinology. 2010;4:1893–901.
Gupta S, Knight AG, Keller JN, Bruce-Keller AJ. Saturated long-chain fatty acids activate inflammatory signaling in astrocytes. J Neurochem. 2012;6:1060–71.
Loikkanen I, Haghighi S, Vainio S, Pajunen A. Expression of cytosolic acetyl-CoA synthetase gene is developmentally regulated. Mech Dev. 2002;1–2:139–41.
Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;2:207–17.
Acknowledgments
This work was supported by NIH R01NS089795 (HTG), NIH R01GM087964 (DCM), and the NIH/NCSU Molecular Biotechnology Training Grant 5T32GM00-8776-08 (PL). FM acknowledges travel funding from DAAD (German Academic Exchange Service).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflicts of interest. Mice used in this study were bred and housed in the College of Veterinary Medicine vivarium according to Institutional Animal Care and Use Committee (IACUC), North Carolina State University regulations, and Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals.
Additional information
Philip Loziuk and Florian Meier contributed equally to this work.
Rights and permissions
About this article
Cite this article
Loziuk, P., Meier, F., Johnson, C. et al. TransOmic analysis of forebrain sections in Sp2 conditional knockout embryonic mice using IR-MALDESI imaging of lipids and LC-MS/MS label-free proteomics. Anal Bioanal Chem 408, 3453–3474 (2016). https://doi.org/10.1007/s00216-016-9421-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9421-3