Abstract
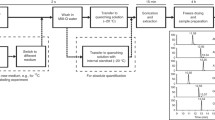
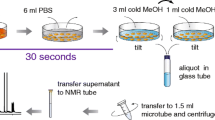
We developed a methodology for the analysis of intracellular metabolites using nuclear magnetic resonance spectrometry (NMR), gas-chromatography coupled with mass spectrometry (GC-MS), and liquid chromatography coupled with high resolution mass spectrometry (LC-HRMS). The main steps for analysis of adherent cells in order to recover the widest possible range of intracellular compounds are blocking metabolic activity by quenching and extraction of intracellular metabolites. We explored three protocols to quench NSC-34 cell metabolism and four different extraction methods, analyzed by NMR. On the basis of the number of metabolites extracted and their relative standard deviation (RSD) analyzed by NMR, the most reproducible protocol [quenching by MeOH at −40 °C and extraction with CH2Cl2/MeOH/H2O (3:3:2)] was used to obtain intracellular media to be analyzed by GC-MS and LC-HRMS. GC-MS analysis was optimized by three oximation procedures followed by silylation derivatization and these were compared to silylation alone. Using reversed-phase liquid chromatography (C18), four different gradients for LC-MS were compared. The analytical protocols were determined to establish the reliability and suitability of sample treatments required to achieve the correct biological analysis of untargeted mammalian cell metabolomics.





Similar content being viewed by others
References
Patti GJ, Yanes O, Siuzdak G (2012) Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol 13(4):263–269
Chrysanthopoulos PK, Goudar CT, Klapa MI (2010) Metabolomics for high-resolution monitoring of the cellular physiological state in cell culture engineering. Metab Eng 12:212–222
Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB (2003) High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol 21:692–696
Wang J-R, Zhang H, Yau LF, Mi J-N, Lee S, Lee KC, Hu P, Liu L, Jiang Z-H (2014) Improved sphingolipidomic approach based on ultra-high performance liquid chromatography and multiple mass spectrometries with application to cellular neurotoxicity. Anal Chem 86:5688–5696
Winder CL, Dunn WB, Schuler S, Broadhurst D, Jarvis R, Stephens GM, Goodacre R (2008) Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites. Anal Chem 80:2939–2948
Bayet-Robert M, Morvan D, Chollet P, Barthomeuf C (2010) Pharmacometabolomics of docetaxel-treated human MCF7 breast cancer cells provides evidence of varying cellular responses at high and low doses. Breast Cancer Res Treat 120:613–626
Cao B, Aa J, Wang G, Wu X, Liu L, Li M, Shi J, Wang X, Zhao C, Zheng T, Guo S, Duan J (2011) GC–TOFMS analysis of metabolites in adherent MDCK cells and a novel strategy for identifying intracellular metabolic markers for use as cell amount indicators in data normalization. Anal Bioanal Chem 400:2983–2993
León Z, García-Cañaveras JC, Donato MT, Lahoz A (2013) Mammalian cell metabolomics: experimental design and sample preparation. Electrophoresis 34:2762–2775
Martineau E, Tea I, Loaëc G, Giraudeau P, Akoka S (2011) Strategy for choosing extraction procedures for NMR-based metabolomic analysis of mammalian cells. Anal Bioanal Chem 401(7):2133–2142
Teng Q, Huang W, Collette T, Ekman D, Tan C (2009) A direct cell quenching method for cell-culture based metabolomics. Metabolomics 5(2):199–208
Lane A, Fan TM (2007) Quantification and identification of isotopomer distributions of metabolites in crude cell extracts using 1H TOCSY. Metabolomics 3(2):79–86
Danielsson AP, Moritz T, Mulder H, Spegel P (2010) Development and optimization of a metabolomic method for analysis of adherent cell cultures. Anal Biochem 404(1):30–39
Sellick CA, Hansen R, Maqsood AR, Dunn WB, Stephens GM, Goodacre R, Dickson AJ (2009) Effective quenching processes for physiologically valid metabolite profiling of suspension cultured mammalian cells. Anal Chem 81(1):174–183
Lorenz MA, Burant CF, Kennedy RT (2011) Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal Chem 83(9):3406–3414
Vuckovic D (2012) Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Anal Bioanal Chem 403(6):1523–1548
Want EJ, Masson P, Michopoulos F, Wilson ID, Theodoridis G, Plumb RS, Shockcor J, Loftus N, Holmes E, Nicholson JK (2013) Global metabolic profiling of animal and human tissues via UPLC-MS. Nat Protoc 8(1):17–32
Naz S, García A, Barbas C (2013) Multiplatform analytical methodology for metabolic fingerprinting of lung tissue. Anal Chem 85(22):10941–10948
Vinaixa M, Rodriguez MA, Samino S, Díaz M, Beltran A, Mallol R, Bladé C, Ibañez L, Correig X, Yanes O (2011) Metabolomics reveals reduction of metabolic oxidation in women with polycystic ovary syndrome after pioglitazone-flutamide-metformin polytherapy. PLoS One 6(12):e29052
Goodpaster AM, Ramadas EH, Kennedy MA (2011) Potential effect of diaper and cotton ball contamination on NMR- and LC/MS-based metabonomics studies of urine from newborn babies. Anal Chem 83(3):896–902
Emond P, Mavel S, Aïdoud N, Nadal-Desbarats L, Montigny F, Bonnet-Brilhault F, Barthélémy C, Merten M, Sarda P, Laumonnier F, Vourc’h P, Blasco H, Andres C (2013) GC-MS-based urine metabolic profiling of autism spectrum disorders. Anal Bioanal Chem 405(15):5291–5300
Nadal-Desbarats L, Aidoud N, Emond P, Blasco H, Filipiak I, Sarda P, Bonnet-Brilhault F, Mavel S, Andres CR (2014) Combined 1H-NMR and 1H-13C HSQC-NMR to improve urinary screening in autism spectrum disorders. Analyst 139(13):3460–3468
Chan EC, Pasikanti KK, Nicholson JK (2011) Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat Protoc 6(10):1483–1499
Mavel S, Nadal-Desbarats L, Blasco H, Bonnet-Brilhault F, Barthelemy C, Montigny F, Sarda P, Laumonnier F, Vourc'h P, Andres CR, Emond P (2013) 1H-13C NMR-based urine metabolic profiling in autism spectrum disorders. Talanta 114:95–102
Martano G, Delmotte N, Kiefer P, Christen P, Kentner D, Bumann D, Vorholt JA (2015) Fast sampling method for mammalian cell metabolic analyses using liquid chromatography–mass spectrometry. Nat Protoc 10(1):1–11
Le Belle JE, Harris NG, Williams SR, Bhakoo KK (2002) A comparison of cell and tissue extraction techniques using high-resolution 1H-NMR spectroscopy. NMR Biomed 15(1):37–44
Dettmer K, Nurnberger N, Kaspar H, Gruber MA, Almstetter MF, Oefner PJ (2011) Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols. Anal Bioanal Chem 399(3):1127–1139
Ritter JB, Genzel Y, Reichl U (2008) Simultaneous extraction of several metabolites of energy metabolism and related substances in mammalian cells: optimization using experimental design. Anal Biochem 373(2):349–369
Dietmair S, Timmins NE, Gray PP, Nielsen LK, Kromer JO (2010) Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal Biochem 404(2):155–164
Loizides-Mangold U (2013) On the future of mass-spectrometry-based lipidomics. FEBS J 280(12):2817–2829
Fan TM, Lane A, Higashi R, Yan J (2011) Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics 7(2):257–269
Čuperlović-Culf M, Barnett DA, Culf AS, Chute I (2010) Cell culture metabolomics: applications and future directions. Drug Discov Today 15:610–621
Li X, Long D, Ji J, Yang W, Zeng Z, Guo S, Ji Z, Qi G, Chen S (2013) Sample preparation for the metabolomics investigation of poly-gamma-glutamate-producing Bacillus licheniformis by GC–MS. J Microbiol Meth 94(1):61–67
Sellick C, Knight D, Croxford A, Maqsood A, Stephens G, Goodacre R, Dickson A (2010) Evaluation of extraction processes for intracellular metabolite profiling of mammalian cells: matching extraction approaches to cell type and metabolite targets. Metabolomics 6(3):427–438
Sana TR, Waddell K, Fischer SM (2008) A sample extraction and chromatographic strategy for increasing LC/MS detection coverage of the erythrocyte metabolome. J Chromatogr B Analyt Technol Biomed Life Sci 871(2):314–321
Cequier-Sanchez E, Rodriguez C, Ravelo AG, Zarate R (2008) Dichloromethane as a solvent for lipid extraction and assessment of lipid classes and fatty acids from samples of different natures. J Agric Food Chem 56(12):4297–4303
Kanani H, Chrysanthopoulos PK, Klapa MI (2008) Standardizing GC–MS metabolomics. J Chromatogr B 871:191–201
Hu X, Li H, Tang P, Sun J, Yuan Q, Li C (2013) GC–MS-based metabolomics study of the responses to arachidonic acid in Blakeslea trispora. Fungal Genet Biol 57:33–41
Gullberg J, Jonsson P, Nordström A, Sjöström M, Moritz T (2004) Design of experiments: an efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal Biochem 331(2):283–295
Koek M, Jellema R, van der Greef J, Tas A, Hankemeier T (2011) Quantitative metabolomics based on gas chromatography mass spectrometry: status and perspectives. Metabolomics 7(3):307–328
Bi H, Krausz KW, Manna SK, Li F, Johnson CH, Gonzalez FJ (2013) Optimization of harvesting, extraction, and analytical protocols for UPLC-ESI-MS-based metabolomic analysis of adherent mammalian cancer cells. Anal Bioanal Chem 405(15):5279–5289
Fei F, Bowdish DME, McCarry BE (2014) Comprehensive and simultaneous coverage of lipid and polar metabolites for endogenous cellular metabolomics using HILIC-TOF-MS. Anal Bioanal Chem 406(15):3723–3733
Gangl ET, Annan M, Spooner N, Vouros P (2001) Reduction of signal suppression effects in ESI-MS using a nanosplitting device. Anal Chem 73(23):5635–5644
Gika HG, Theodoridis GA, Plumb RS, Wilson ID (2014) Current practice of liquid chromatography–mass spectrometry in metabolomics and metabonomics. J Pharm Biomed 87:12–25
Paglia G, Hrafnsdóttir S, Magnúsdóttir M, Fleming RT, Thorlacius S, Palsson B, Thiele I (2012) Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole–time of flight mass spectrometry (UPLC–Q–ToF-MS). Anal Bioanal Chem 402(3):1183–1198
Abdel Rahman AM, Pawling J, Ryczko M, Caudy AA, Dennis JW (2014) Targeted metabolomics in cultured cells and tissues by mass spectrometry: method development and validation. Anal Chim Acta 845:53–61
Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD (2010) Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem 82(8):3212–3221
Acknowledgments
We thank the “Département d’Analyses Chimiques et S.R.M. Biologique et Médicale” (PPF, Tours, France) for chemical analyses and especially Cinzia Bocca for LC-HRMS analyses.
Funding
This study was funded by ARSLA (“Association pour la Recherche sur la Sclérose Latérale Amyotrophique et autres maladies du motoneurone”) with a research grant and by “La Région Centre” with a PhD graduate grant. This work was supported by the “Institut National de la Santé et de la Recherche” INSERM and the University François-Rabelais.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 719 kb)
Rights and permissions
About this article
Cite this article
Madji Hounoum, B., Blasco, H., Nadal-Desbarats, L. et al. Analytical methodology for metabolomics study of adherent mammalian cells using NMR, GC-MS and LC-HRMS. Anal Bioanal Chem 407, 8861–8872 (2015). https://doi.org/10.1007/s00216-015-9047-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-015-9047-x