Abstract
Metabolomic results on human blood plasma largely depend on the sample preparation protocols employed for protein precipitation and metabolite extraction. Five different extraction methods were examined, which can be grouped into two categories, liquid-liquid extraction and protein precipitation methods, including long-standing protocols such as the Folch extraction and Bligh-Dyer extraction in comparison to modern methods such as the Matyash protocol and two global metabolite extraction methods. Extracts were subjected to analysis of blood plasma lipids and primary metabolites by using chip-based direct infusion nanoelectrospray tandem mass spectrometry and gas chromatography coupled to time-of-flight mass spectrometry, respectively. Optimal extraction schemes were evaluated based on the number of identified metabolites, extraction efficiency, compound diversity, reproducibility, and convenience for high-throughput sample preparations. Results showed that Folch and Matyash methods were equally valid and robust for lipidomic assessments while primary metabolites were better assessed by the protein precipitation methods with organic solvent mixtures.

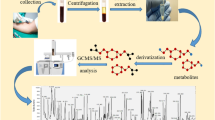
Schematic workflow of five extraction methods and subsequent mass spectrometry analysis using GC-TOF MS and nanoelectrospray direct-infusion ion trap MS/MSᅟ






Similar content being viewed by others
References
Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B (2011) The human serum metabolome. PLoS ONE 6(2):e16957
Serkova NJ, Standiford TJ, Stringer KA (2011) The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Respir Crit Care Med 184(6):647
Griffiths WJ, Wang Y (2009) Mass spectrometry: from proteomics to metabolomics and lipidomics. Chem Soc Rev 38(7):1882–1896
Fiehn O (2001) Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comparative Funct Genomics 2(3):155–168
Crews B, Wikoff WR, Patti GJ, Woo H-K, Kalisiak E, Heideker J, Siuzdak G (2009) Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Anal Chem 81(20):8538–8544
Bogdanov M, Matson WR, Wang L, Matson T, Saunders-Pullman R, Bressman SS, Beal MF (2008) Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain 131(2):389–396
Orešič M, Hyötyläinen T, Herukka S, Sysi-Aho M, Mattila I, Seppänan-Laakso T, Julkunen V, Gopalacharyulu P, Hallikainen M, Koikkalainen J (2011) Metabolome in progression to Alzheimer’s disease. Translatl Psychiatry 1(12):e57
Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, Heim K, Campillos M, Holzapfel C, Thorand B (2012) Novel biomarkers for pre-diabetes identified by metabolomics. Molecular systems biology 8:615
Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A (2006) Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal Chem 78(2):585–595
Gao X, Zhang Q, Meng D, Isaac G, Zhao R, Fillmore TL, Chu RK, Zhou J, Tang K, Hu Z (2012) A reversed-phase capillary ultra-performance liquid chromatography–mass spectrometry (UPLC-MS) method for comprehensive top-down/bottom-up lipid profiling. Anal Bioanal Chem 402(9):2923–2933
Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL (2010) Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res 51(11):3299–3305
Sandra K, Pereira AS, Vanhoenacker G, David F, Sandra P (2010) Comprehensive blood plasma lipidomics by liquid chromatography/quadrupole time-of-flight mass spectrometry. J Chromatogr A 1217(25):4087–4099
Nie H, Liu R, Yang Y, Bai Y, Guan Y, Qian D, Wang T, Liu H (2010) Lipid profiling of rat peritoneal surface layers by online normal-and reversed-phase 2D LC QToF-MS. J Lipid Res 51(9):2833–2844
Laaksonen R, Katajamaa M, Päivä H, Sysi-Aho M, Saarinen L, Junni P, Lütjohann D, Smet J, Van Coster R, Seppänen-Laakso T (2006) A systems biology strategy reveals biological pathways and plasma biomarker candidates for potentially toxic statin-induced changes in muscle. PLoS ONE 1(1):e97
Pizarro C, Arenzana-Rámila I, Pérez-del-Notario N, Pérez-Matute P, González-Sáiz J-M (2013) Plasma lipidomic profiling method based on ultrasound extraction and liquid chromatography mass spectrometry. Anal Chem 85(24):12085–12092. doi:10.1021/ac403181c
Ståhlman M, Ejsing CS, Tarasov K, Perman J, Borén J, Ekroos K (2009) High-throughput shotgun lipidomics by quadrupole time-of-flight mass spectrometry. J Chromatogr B 877(26):2664–2672
Han X, Gross RW (2005) Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev 24(3):367–412
Han X, Gross RW (2003) Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry a bridge to lipidomics. J Lipid Res 44(6):1071–1079
Han X, Yang K, Gross RW (2012) Multidimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev 31(1):134–178
Graessler J, Schwudke D, Schwarz PE, Herzog R, Shevchenko A, Bornstein SR (2009) Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS ONE 4(7):e6261
Lin L, Yu Q, Yan X, Hang W, Zheng J, Xing J, Huang B (2010) Direct infusion mass spectrometry or liquid chromatography mass spectrometry for human metabonomics? A serum metabonomic study of kidney cancer. Analyst 135(11):2970–2978
Thomas A, Déglon J, Lenglet S, Mach F, Mangin P, Wolfender J-L, Steffens S, Staub C (2010) High-throughput phospholipidic fingerprinting by online desorption of dried spots and quadrupole-linear ion trap mass spectrometry: evaluation of atherosclerosis biomarkers in mouse plasma. Anal Chem 82(15):6687–6694
Li F, Qin X, Chen H, Qiu L, Guo Y, Liu H, Chen G, Song G, Wang X, Li F (2013) Lipid profiling for early diagnosis and progression of colorectal cancer using direct infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom 27(1):24–34
Yang L, Bennett R, Strum J, Ellsworth BB, Hamilton D, Tomlinson M, Wolf RW, Housley M, Roberts BA, Welsh J (2009) Screening phosphatidylcholine biomarkers in mouse liver extracts from a hypercholesterolemia study using ESI-MS and chemometrics. Anal Bioanal Chem 393(2):643–654
Rappley I, Myers DS, Milne SB, Ivanova PT, LaVoie MJ, Brown HA, Selkoe DJ (2009) Lipidomic profiling in mouse brain reveals differences between ages and genders, with smaller changes associated with α-synuclein genotype. J Neurochem 111(1):15–25
Shen Q, Wang Y, Gong L, Guo R, Dong W, Cheung H-Y (2012) Shotgun lipidomics strategy for fast analysis of phospholipids in fisheries waste and its potential in species differentiation. J Agric Food Chem 60(37):9384–9393
Vu HS, Tamura P, Galeva NA, Chaturvedi R, Roth MR, Williams TD, Wang X, Shah J, Welti R (2012) Direct infusion mass spectrometry of oxylipin-containing Arabidopsis membrane lipids reveals varied patterns in different stress responses. Plant Physiol 158(1):324–339
Raterink R-J, van der Kloet FM, Li J, Wattel NA, Schaaf MJM, Spaink HP, Berger R, Vreeken RJ, Hankemeier T (2013) Rapid metabolic screening of early zebrafish embryogenesis based on direct infusion-nanoESI-FTMS. Metabolomics 9(4):864–873
Holguin FO, Schaub T (2013) Characterization of microalgal lipid feedstock by direct-infusion FT-ICR mass spectrometry. Algal Res 2(1):43–50
Basconcillo LS, Zaheer R, Finan TM, McCarry BE (2009) A shotgun lipidomics approach in Sinorhizobium meliloti as a tool in functional genomics. J Lipid Res 50(6):1120–1132
Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D (2008) Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 49(5):1137–1146
Schuhmann K, Almeida R, Baumert M, Herzog R, Bornstein SR, Shevchenko A (2012) Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J Mass Spectrom 47(1):96–104
Fiehn O, Garvey WT, Newman JW, Lok KH, Hoppel CL, Adams SH (2010) Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS ONE 5(12):e15234
Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y (2009) Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457(7231):910–914
Simón-Manso Y, Lowenthal MS, Kilpatrick LE, Sampson ML, Telu KH, Rudnick PA, Mallard WG, Bearden DW, Schock TB, Tchekhovskoi DV (2013) Metabolite profiling of a NIST standard reference material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal Chem 85(24):11725–11731
Ismaiel OA, Zhang T, Jenkins RG, Karnes HT (2010) Investigation of endogenous blood plasma phospholipids, cholesterol and glycerides that contribute to matrix effects in bioanalysis by liquid chromatography/mass spectrometry. J Chromatogr B 878(31):3303–3316
Reis A, Rudnitskaya A, Blackburn GJ, Fauzi NM, Pitt AR, Spickett CM (2013) A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J Lipid Res 54(7):1812–1824
Trygg J, Gullberg J, Johansson AI, Jonsson P, Antti H, Marklund SL, Moritz T (2005) Extraction and GC/MS analysis of the human blood plasma metabolome. Anal Chem 77(24):8086–8094
Bruce SJ, Tavazzi I, Parisod V, Rezzi S, Kochhar S, Guy PA (2009) Investigation of human blood plasma sample preparation for performing metabolomics using ultrahigh performance liquid chromatography/mass spectrometry. Anal Chem 81(9):3285–3296
Bruce SJ, Jonsson P, Antti H, Cloarec O, Trygg J, Marklund SL, Moritz T (2008) Evaluation of a protocol for metabolic profiling studies on human blood plasma by combined ultra-performance liquid chromatography/mass spectrometry: from extraction to data analysis. Anal Biochem 372(2):237–249
Fiehn O, Wohlgemuth G, Scholz M Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. In, 2005. Springer, pp 735-735
Kind T, Liu K-H, Lee DY, DeFelice B, Meissen JK, Fiehn O (2013) LipidBlast in silico tandem mass spectrometry database for lipid identification. Nature Methods
Folch J, Lees M, Sloane-Stanley G (1957) A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226(1):497–509
Bligh E, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37(8):911–917
Fiehn O, Kind T (2007) Metabolite profiling in blood plasma. Metabolomics. Springer, In, pp 3–17
Lee DY, Fiehn O (2008) High quality metabolomic data for Chlamydomonas reinhardtii. Plant Methods 4(1):7
Mayampurath AM, Jaitly N, Purvine SO, Monroe ME, Auberry KJ, Adkins JN, Smith RD (2008) DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics 24(7):1021–1023
Frank AM, Bandeira N, Shen Z, Tanner S, Briggs SP, Smith RD, Pevzner PA (2007) Clustering millions of tandem mass spectra. J Proteome Res 7(01):113–122
Stein SE, Scott DR (1994) Optimization and testing of mass spectral library search algorithms for compound identification. J Am Soc Mass Spectrom 5(9):859–866
Song I-S, Lee DY, Shin M-H, Kim H, Ahn YG, Park I, Kim KH, Kind T, Shin J-G, Fiehn O, Liu K-H (2012) Pharmacogenetics meets metabolomics: discovery of tryptophan as a new endogenous oct2 substrate related to metformin disposition. PLoS ONE 7(5):e36637
Kind T, Tolstikov V, Fiehn O, Weiss RH (2007) A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal Biochem 363(2):185–195
Scholz M, Fiehn O SetupX–a public study design database for metabolomic projects. In, 2007. Pac Symp Biocomput, pp 169–180
Saeed A, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34(2):374
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95(25):14863–14868
Taylor PJ (2005) Matrix effects: the Achilles heel of quantitative high-performance liquid chromatography–electrospray–tandem mass spectrometry. Clin Biochem 38(4):328–334
Kim S, Lee DY, Wohlgemuth G, Park HS, Fiehn O, Kim KH (2013) Evaluation and optimization of metabolome sample preparation methods for Saccharomyces cerevisiae. Anal Chem 85(4):2169–2176
Hutchins PM, Barkley RM, Murphy RC (2008) Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J Lipid Res 49(4):804–813
Liu L, Aa J, Wang G, Yan B, Zhang Y, Wang X, Zhao C, Cao B, Shi J, Li M (2010) Differences in metabolite profile between blood plasma and serum. Anal Biochem 406(2):105–112
Begley P, Francis-McIntyre S, Dunn WB, Broadhurst DI, Halsall A, Tseng A, Knowles J, Goodacre R, Kell DB (2009) Development and performance of a gas chromatography−time-of-flight mass spectrometry analysis for large-scale nontargeted metabolomic studies of human serum. Anal Chem 81(16):7038–7046
Acknowledgments
This study was supported by the National Institutes of Health grants 1U24 DK097154 and P20 HL113452 to OF, the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning (grant no. NRF-2013M3A9B6046416), the Korea Healthcare Technology R&D Project funded by Ministry of Health & Welfare (grant no. HN13C0076), and the Advanced Biomass R&D Center of Korea funded by the Korean Government (MSIP, grant no. 2011-0031353), Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 5501 kb)
Rights and permissions
About this article
Cite this article
Lee, D.Y., Kind, T., Yoon, YR. et al. Comparative evaluation of extraction methods for simultaneous mass-spectrometric analysis of complex lipids and primary metabolites from human blood plasma. Anal Bioanal Chem 406, 7275–7286 (2014). https://doi.org/10.1007/s00216-014-8124-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-8124-x