Abstract
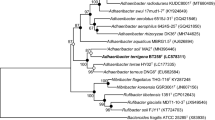
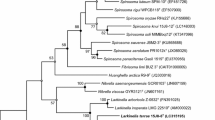
A Gram-stain-negative, aerobic, non-motile and yellow-colored bacterium, strain 17J57-3 T, was isolated from soil collected in Pyeongchang city, Korea. Phylogenetic analyses based on 16S rRNA gene sequences revealed that strain 17J57-3 T formed a distinct lineage within the family Oxalobacteraceae (order Burkholderiales, class Betaproteobacteria). Strain 17J57-3 T was the most closely related to Noviherbaspirillum humi U15T (96.4% 16S rRNA gene sequence similarity) and Noviherbaspirillum massiliense JC206T (96.2%). The draft genome size of strain 17J57-3 T was 6,117,206 bp. Optimal growth occurred at 30 °C, pH 7.0 without NaCl. The predominant cellular fatty acids were summed feature 3 (C16:1 ω6c/C16:1 ω7c) and C16:0. The major respiratory quinone was Q-8. The major polar lipids were diphosphatidylglycerol, phosphatidylglycerol and phosphatidylethanolamine. Biochemical, chemotaxonomic and phylogenetic analyses indicated that strain 17J57-3 T represents a novel bacterial species within the genus Noviherbaspirillum, for which the name Noviherbaspirillum galbum is proposed. The type strain of Noviherbaspirillum galbum is 17J57-3 T (= KCTC 62213 T = NBRC 114384 T).

Similar content being viewed by others
References
Cappuccino JG, Sherman N (2002) Microbiology—a laboratory manual, 6th edn. Pearson Education, Inc. Benjamin Cummings, California
Carro L, Rivas R, León-Barrios M, González-Tirante M, Velázquez E, Valverde A (2012) Herbaspirillum canariense sp. nov., Herbaspirillum aurantiacum sp. nov. and Herbaspirillum soli sp. nov., isolated from volcanic mountain soil, and emended description of the genus Herbaspirillum. Int J Syst Evol Microbiol 62:1300–1306. https://doi.org/10.1099/ijs.0.031336-0
Chaudhary DK, Kim J (2017) Noviherbaspirillum agri sp. nov., isolated from reclaimed grassland soil, and reclassification of Herbaspirillum massiliense (Lagier et al. 2014) as Noviherbaspirillum massiliense comb. nov. Int J Syst Evol Microbiol 67:1508–1515. https://doi.org/10.1099/ijsem.0.001747
de Boer W, Leveau JHJ, Kowalchuk GA, Klein Gunnewiek PJA, Abeln ECA, Figge MJ, Sjollema K, Janse JD, van Veen JA (2004) Collimonas fungivorans gen. nov., sp. nov., a chitinolytic soil bacterium with the ability to grow on living fungal hyphae. Int J Syst Evol Microbiol 54(3):857–864
Dobritsa AP, Reddy MCS, Samadpour M (2010) Reclassification of Herbaspirillum putei as a later heterotypic synonym of Herbaspirillum huttiense, with the description of H. huttiense subsp. huttiense subsp. nov. and H. huttiense subsp. putei subsp. nov., comb. nov., and description of Herbaspirillum aquaticum sp. nov.. Int J Syst Evol Microbiol 60(6):1418–1426
Fan MC, Guo YQ, Zhang LP et al (2018) Herbaspirillum robiniae sp. nov., isolated from root nodules of Robinia pseudoacacia in a lead-zinc mine. Int J Syst Evol Microbiol 68:1300–1306
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17(6):368–376
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20(4):406
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Höppener-Ogawa S, de Boer W, Leveau JH et al (2008) Collimonas arenae sp. nov. and Collimonas pratensis sp. nov., isolated from (semi-)natural grassland soils. Int J Syst Evol Microbiol 58:414–419
Im WT, Bae HS, Yokota A, Lee ST (2004) Herbaspirillum chlorophenolicum sp. nov., a 4-chlorophenol-degrading bacterium. Int J Syst Evol Microbiol 2004:851–855
Ishii S, Ashida N, Ohno H et al (2017) Noviherbaspirillum denitrificans sp. nov., a denitrifying bacterium isolated from rice paddy soil and Noviherbaspirillum autotrophicum sp. nov., a denitrifying, facultatively autotrophic bacterium isolated from rice paddy soil and proposal to reclassify Herbaspirillum massiliense as Noviherbaspirillum massiliense comb. nov. Int J Syst Evol Microbiol 67:1841–1848
Jung S-Y, Lee M-H, Oh T-K, Yoon J-H (2007) Herbaspirillum rhizosphaerae sp. nov., isolated from rhizosphere soil of Allium victorialis var. platyphyllum. Int J Syst Evol Microbiol 57(10):2284–2288
Kim SJ, Moon JY, Weon HY, Hong SB, Seok SJ, Kwon SW (2014a) Noviherbaspirillum suwonense sp. nov., isolated from an air sample. Int J Syst Evol Microbiol 64:1552–1558. https://doi.org/10.1099/ijs.0.057372-0
Kim SJ, Moon JY, Weon HY, Hong SB, Seok SJ, Kwon SW (2014b) Noviherbaspirillum suwonense sp. Nov., isolated from an air sample. Int J Syst Evol Microbiol 64(Pt 5):1552–1558. https://doi.org/10.1099/ijs.0.057372-0
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Kirchhof G, Eckert B, Stoffels M, Baldani JI, Reis VM, Hartmann A (2001) Herbaspirillum frisingense sp. nov., a new nitrogen-fixing bacterial species that occurs in C4-fibre plants. Int J Syst Evol Microbiol 51:157–168
Komagata K, Suzuki K (1988) 4 Lipid and cell-wall analysis in bacterial systematics. Method Microbiol 19:161–207
Konstantinidis KT, Tiedje JM (2005) Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102:2567–2572
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Lee SD (2018) Collimonas antrihumi sp. nov., isolated from a natural cave and emended description of the genus Collimonas. Int J Syst Evol Microbiol 68:2448–2453
Lee I, Kim YO, Park CJ (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Lin SY, Hameed A, Arun AB, Liu YC, Hsu YH, Lai WA, Rekha PD, Young CC (2013) Description of Noviherbaspirillum malthae gen. nov., sp. nov., isolated from an oil-contaminated soil, and proposal to reclassify Herbaspirillum soli, Herbaspirillum aurantiacum, Herbaspirillum canariense and Herbaspirillum psychrotolerans as Noviherbaspirillum soli comb. nov., Noviherbaspirillum aurantiacum comb. nov., Noviherbaspirillum canariense comb. nov. and Noviherbaspirillum psychrotolerans comb. nov. based on polyphasic analysis. Int J Syst Evol Microbiol 63:4100–4107. https://doi.org/10.1099/ijs.0.048231-0
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence–based species delimitation with confidence intervals and improved distance functions. BMC Bioinf 14:60
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Meth 2:233–241
Rothballer M, Schmid M, Klein I, Gattinger A, Grundmann S, Hartmann A (2006) Herbaspirillum hiltneri sp. nov., isolated from surface-sterilized wheat roots. Int J Syst Evol Microbiol 56(6):1341–1348
Sahin N, Portillo MC, Kato Y, Schumann P (2009) Description of Oxalicibacterium horti sp. nov. and Oxalicibacterium faecigallinarum sp. nov., new aerobic, yellow-pigmented, oxalotrophic bacteria. FEMS Microbiol Lett 296(2):198–202
Sahin N, Gonzalez JM, Iizuka T, Hill JE (2010) Characterization of two aerobic ultramicrobacteria isolated from urban soil and a description of Oxalicibacterium solurbis sp. nov. FEMS Microbiol Lett 307:25–29. https://doi.org/10.1111/j.1574-6968.2010.01954.x
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc, Newark
Smibert RM, Krieg NR (1981) General characterization. Manual of methods for general bacteriology. American Society for Microbiology, Washington, pp 409–442
Sundararaman A, Srinivasan S, Lee SS (2016) Noviherbaspirillum humi sp. nov., isolated from soil. Antonie Van Leeuwenhoek 109:697–704. https://doi.org/10.1007/s10482-016-0670-0
Tamer AÜ, Aragno M, Şahin N (2002) Isolation and characterization of a new type of aerobic, oxalic acid utilizing bacteria, and proposal of Oxalicibacterium flavum gen. nov., sp. nov.. Syst Appl Microbiol 25(4):513–519
Tatusova T, DiCuccio M, Badretdin A et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. https://doi.org/10.1093/nar/gkw569
Weisburg WG, Barns SM, Pellerier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Xu G, Chang J, Xue H, Guo M, Piao CG, Li Y (2018) Herbaspirillum piri sp. nov., isolated from bark of a pear tree. Int J Syst Evol Microbiol 68:3652–3656
Acknowledgements
This work was supported by a research grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR201902203). This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01505201)” Rural Development Administration, Republic of Korea. This research was supported by the MISP (Ministry of Science, ICT and Future Planning), Korea, under the National Program for Excellence in SW) (2016-0-00022) supervised by the IITP(Institute of Information and communications Technology Planning and Evaluation). We are grateful to Dr. Aharon Oren (The Hebrew University of Jerusalem, Israel) for helping with the etymology. The GenBank accession numbers for the 16S rRNA gene sequences of strain 17J57-3T is MN396447. The whole-genome sequences of strain 17J57-3T have been deposited into DDBJ/EMBL/GenBank under the accession numbers JAAIVB000000000.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Park, Y., Maeng, S., Lee, S.E. et al. Noviherbaspirillum galbum sp. nov., a bacterium isolated from soil. Arch Microbiol 203, 823–828 (2021). https://doi.org/10.1007/s00203-020-02076-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-02076-7