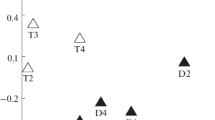
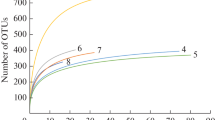
Abstract
The bacterioplankton diversity of coastal waters along a latitudinal gradient between Puerto Rico and Argentina was analyzed using a total of 134,197 high-quality sequences from the V6 hypervariable region of the small-subunit ribosomal RNA gene (16S rRNA) (mean length of 60 nt). Most of the OTUs were identified into Proteobacteria, Bacteriodetes, Cyanobacteria, and Actinobacteria, corresponding to approx. 80% of the total number of sequences. The number of OTUs corresponding to species varied between 937 and 1946 in the seven locations. Proteobacteria appeared at high frequency in the seven locations. An enrichment of Cyanobacteria was observed in Puerto Rico, whereas an enrichment of Bacteroidetes was detected in the Argentinian shelf and Uruguayan coastal lagoons. The highest number of sequences of Actinobacteria and Acidobacteria were obtained in the Amazon estuary mouth. The rarefaction curves and Good coverage estimator for species diversity suggested a significant coverage, with values ranging between 92 and 97% for Good coverage. Conserved taxa corresponded to aprox. 52% of all sequences. This study suggests that human-contaminated environments may influence bacterioplankton diversity.




Similar content being viewed by others
References
Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM (2009) A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4:6372
Andersson AF, Riemann L, Bertilsson S (2010) Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J 4:171–181
Andrade L, Gonzalez AM, Araujo FV, Paranhos R (2003) Flow cytometry assessment of bacterioplankton in tropical marine environments. J Microbiol Methods 19:89–94
Artigas LF, Otero E, Paranhos R, Gomez ML, Piccini C, Costagliola M, Silva R, Suarez P, Gallardo VA, Hernandez Becerril DU, Chistoserdov A, Vieira R, Perez-Cenci M, Ternon JF, Beber B, Thyssen M, Dionisi H, do Rosario Marinho-Jassaud I, Gonzalez A, Hurtado C, Parra JP, Alonso C, Hozbor C, Peresutti S, Negri R, Espinoza C, Cardoso A, Martins O, Covacevich F, Berón C, Salerno GL (2008) Towards a Latin American and Caribbean international census of marine microbes (LACar-ICoMM): overview and discussion on some current research directions. Int J TropBiol Conserv 56:183–214
Brown MV, Philip GK, Bunge JA, Smith MC, Bissett A, Lauro FM, Fuhrman JA, Donachie SP (2009) Microbial community structure in the North Pacific Ocean. ISME J 3:1286–1300
Castro AP, Araújo SD Jr, Reis AM, Moura RL, Francini-Filho RB, Pappas G Jr, Rodrigues TB, Thompson FL, Krüger RH (2010) Bacterial community associated with healthy and diseased reef coral Mussismilia hispida from eastern Brazil. Microb Ecol 59:658–667
Claesson MJ, O’Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O’Toole PW (2009) Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4(8):e6669
Demba Diallo M, Willems A, Vloemans N, Cousin S, Vandekerckhove TT, de Lajudie P, Neyra M, Vyverman W, Gillis M, Van der Gucht K (2004) Polymerase chain reaction denaturing gradient gel electrophoresis analysis of the N2-fixing bacterial diversity in soil under Acacia tortilis ssp. raddiana and Balanites aegyptiaca in the dryland part of Senegal. Environ Microbiol 6:400–415
Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, Hatay M, Hall D, Brown E, Haynes M, Krause L, Sala E, Sandin SA, Thurber RV, Willis BL, Azam F, Knowlton N, Rohwer F (2008) Microbial ecology of four coral atolls in the Northern Line Islands. PLoS One 3:1584
Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, Brown JH (2008) A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA 105:7774–7778
Galand PE, Casamayor EO, Kirchman DL, Lovejoy C (2009) Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA 106:22427–22432
Gasol JM, del Giorgio PA (2000) Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci Mar 64:197–224
Grasshoff K, Kremling K, Erhardt M (1999) Methods of seawater analysis, 3rd edn. Wiley-VCH Verlag, New York
Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML (2007) Microbial Population Structures in the Deep Marine Biosphere. Science 318:97–100
Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML (2008) Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genetics 4:255
Lins-de-Barros MM, Vieira RP, Cardoso AM, Monteiro VA, Turque AS, Silveira CB, Albano RM, Clementino MM, Martins OB (2010) Archaea, Bacteria, and algal plastids associated with the reef-building corals Siderastrea stellata and Mussismilia hispida from Búzios, South Atlantic Ocean, Brazil. Microb Ecol 59:523–532
Kirchman DL (2008) Microbial ecology of the oceans, 2nd Edition. 620 p. Wiley
May AC (2004) Percent sequence identity; the need to be explicit. Structure 12:737–738
Morris RM, Rappé MS, Connon SA, Vergin KL, Carlson CA, Giovannoni SJ (2002) SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48:443–453
O’Neill B, Grossman J, Tsai MT, Gomes JE, Lehmann J, Peterson J, Neves E, Thies JE (2009) Bacterial community composition in Brazilian Anthrosols and adjacent soils characterized using culturing and molecular identification. Microb Ecol 58:23–35
Pedrós-Alió C (2006) Marine microbial diversity: can it be determined? Trends Microbiol 14:257–263
Piccini C, Conde D, Alonso C, Sommaruga R, Pernthaler J (2006) Blooms of single bacterial species in a coastal lagoon of the southwestern Atlantic Ocean. Appl Environ Microbiol 72:6560–6568
Porter KG, Feig YS (1980) The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25:943–948
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6:639–641
Reis AM, Araújo SD Jr, Moura RL, Francini-Filho RB, Pappas G Jr, Coelho AM, Krüger RH, Thompson FL (2009) Bacterial diversity associated with the Brazilian endemic reef coral Mussismilia braziliensis. J Appl Microbiol 106:1378–1387
Rocap G, Distel DL, Waterbury JB, Chisholm SW (2002) Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S–23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol 68(3):1180–11891
Schauer R, Bienhold C, Ramette A, Harder J (2010) Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J 4:159–170
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA 103:12115–12120
Strateva T, Yordanov D (2009) Pseudomonas aeruginosa—a phenomenon of bacterial resistance. J Med Microbiol 58:1133–1148
Towner KJ (2009) Acinetobacter: an old friend, but a new enemy. J Hosp Infect 73:355–363
Van Trappen S, Mergaert J, Van Eygen S, Dawyndt P, Cnockaert MC, Swings J (2002) Diversity of 746 heterotrophic bacteria isolated from microbial mats from tem Antarctic lakes. Syst Appl Microbiol 4:603–610
Vieira RP, Gonzalez AM, Cardoso AM, Oliveira DN, Albano RM, Clementino MM, Martins OB, Paranhos R (2008) Relationships between bacterial diversity and environmental variables in a tropical marine environment, Rio de Janeiro. Environ Microbiol 10:189–199
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Acknowledgments
Tag sequencing was kindly provided by MBL-WHOI, based on a Keck Foundation grant to M. Sogin & L. Amaral-Zetler. This is a contribution of the Latin American and Caribbean network (LACar-ICoMM) to the International Census of Marine Microbes (ICoMM). Also thanks to NOAA Grant (NA06NOS4780190) for providing support to E. Otero (Puerto Rico) in the form of materials and travel. Thanks are due to the CPAq-IEPA team for its technical and logistical support in conducting the sampling and chemical analysis in the Amazon estuary, funded by the GERCO program (State of Amapa, Brazil). Thanks are due to the French EC2CO CARBAMA program conducted by G. Abril that supported L.F. Artigas and to J. Chicheportiche for help in bacterial and chlorophyll analysis. FLT thanks CNPq, FAPERJ, and IFS for grants. TB thanks CNPq for a PhD scholarship. Guanabara Bay Microbial Observatory and sampling funded by CNPq, and AG thank CNPq for her post-doc grant. We also thank PEDECIBA in Uruguay for supporting the sampling.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thompson, F.L., Bruce, T., Gonzalez, A. et al. Coastal bacterioplankton community diversity along a latitudinal gradient in Latin America by means of V6 tag pyrosequencing. Arch Microbiol 193, 105–114 (2011). https://doi.org/10.1007/s00203-010-0644-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-010-0644-y