Abstract
Summary
Aiming to identify pleiotropic genomic loci for bone mineral density and bone size, we performed a bivariate GWAS in five discovery samples and replicated in two large-scale samples. We identified 2 novel loci at 2q37.1 and 6q26. Our findings provide insight into common genetic architecture underlying both traits.
Introduction
Bone mineral density (BMD) and bone size (BS) are two important factors that contribute to the development of osteoporosis and osteoporotic fracture. Both BMD and BS are highly heritable and they are genetically correlated. In this study, we aim to identify pleiotropic loci associated with BMD and BS.
Methods
We conducted a bivariate genome-wide association (GWA) analysis of hip BMD and hip BS in 6180 participants from 5 samples, followed by in silico replication in the UK Biobank study of BMD (N = 426,824) and the deCODE study of BS (N = 28,954), respectively.
Results
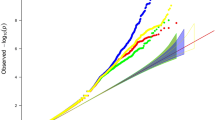
SNPs from 2 genomic loci were significant at the genome-wide significance (GWS) level (p lt; 5 × 10−8) in the discovery samples and were successfully replicated in the replication samples (2q37.1, lead SNP rs7575512, discovery p = 1.49 × 10−10, replication p = 0.05; 6q26, lead SNP rs1040724, discovery p = 1.95 × 10−8, replication p = 0.03). Functional annotations suggested functional relevance of the identified variants to bone development.
Conclusion
Our findings provide insight into the common genetic architecture underlying BMD and BS, and enhance our understanding of the potential mechanism of osteoporosis fracture.




Similar content being viewed by others
References
Eisman JA (1999) Genetics of osteoporosis. Endocr Rev 20(6):788–804. https://doi.org/10.1210/edrv.20.6.0384
Deng HW, Deng XT, Conway T, Xu FH, Heaney R, Recker RR (2002) Determination of bone size of hip, spine, and wrist in human pedigrees by genetic and lifestyle factors. J Clin Densitom 5(1):45–56. https://doi.org/10.1385/jcd:5:1:045
Ahlborg HG, Johnell O, Turner CH, Rannevik G, Karlsson MK (2003) Bone loss and bone size after menopause. N Engl J Med 349(4):327–334. https://doi.org/10.1056/NEJMoa022464
Tan LJ, Liu YZ, Xiao P, Yang F, Tang ZH, Liu PY, Recker RR, Deng HW (2008) Evidence for major pleiotropic effects on bone size variation from a principal component analysis of 451 Caucasian families. Acta Pharmacol Sin 29(6):745–751. https://doi.org/10.1111/j.1745-7254.2008.00806.x
Havill LM, Mahaney MC, T LB, Specker BL (2007) Effects of genes, sex, age, and activity on BMC, bone size, and areal and volumetric BMD. J Bone Miner Res 22(5):737–746. https://doi.org/10.1359/jbmr.070213
Liu YZ, Wilson SG, Wang L, Liu XG, Guo YF, Li J, Yan H, Deloukas P, Soranzo N, Chinappen-Horsley U, Cervino A, Williams FM, Xiong DH, Zhang YP, Jin TB, Levy S, Papasian CJ, Drees BM, Hamilton JJ, Recker RR, Spector TD, Deng HW (2008) Identification of PLCL1 gene for hip bone size variation in females in a genome-wide association study. PLoS One 3(9):e3160. https://doi.org/10.1371/journal.pone.0003160
Deng FY, Dong SS, Xu XH, Liu YJ, Liu YZ, Shen H, Tian Q, Li J, Deng HW (2013) Genome-wide association study identified UQCC locus for spine bone size in humans. Bone 53(1):129–133. https://doi.org/10.1016/j.bone.2012.11.028
Zhang L, Choi HJ, Estrada K, Leo PJ, Li J, Pei YF, Zhang Y, Lin Y, Shen H, Liu YZ, Liu Y, Zhao Y, Zhang JG, Tian Q, Wang YP, Han Y, Ran S, Hai R, Zhu XZ, Wu S, Yan H, Liu X, Yang TL, Guo Y, Zhang F, Guo YF, Chen Y, Chen X, Tan L, Zhang L, Deng FY, Deng H, Rivadeneira F, Duncan EL, Lee JY, Han BG, Cho NH, Nicholson GC, McCloskey E, Eastell R, Prince RL, Eisman JA, Jones G, Reid IR, Sambrook PN, Dennison EM, Danoy P, Yerges-Armstrong LM, Streeten EA, Hu T, Xiang S, Papasian CJ, Brown MA, Shin CS, Uitterlinden AG, Deng HW (2014) Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet 23(7):1923–1933. https://doi.org/10.1093/hmg/ddt575
Kemp JP, Morris JA, Medina-Gomez C, Forgetta V, Warrington NM, Youlten SE, Zheng J, Gregson CL, Grundberg E, Trajanoska K, Logan JG, Pollard AS, Sparkes PC, Ghirardello EJ, Allen R, Leitch VD, Butterfield NC, Komla-Ebri D, Adoum AT, Curry KF, White JK, Kussy F, Greenlaw KM, Xu C, Harvey NC, Cooper C, Adams DJ, Greenwood CMT, Maurano MT, Kaptoge S, Rivadeneira F, Tobias JH, Croucher PI, Ackert-Bicknell CL, Bassett JHD, Williams GR, Richards JB, Evans DM (2017) Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat Genet 49(10):1468–1475. https://doi.org/10.1038/ng.3949
Kim SK (2018) Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS One 13(7):e0200785. https://doi.org/10.1371/journal.pone.0200785
Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai RC, Vulpescu NA, Forgetta V, Kleinman A, Mohanty ST, Sergio CM, Quinn J, Nguyen-Yamamoto L, Luco AL, Vijay J, Simon MM, Pramatarova A, Medina-Gomez C, Trajanoska K, Ghirardello EJ, Butterfield NC, Curry KF, Leitch VD, Sparkes PC, Adoum AT, Mannan NS, Komla-Ebri DSK, Pollard AS, Dewhurst HF, Hassall TAD, Beltejar MG, Me Research T, Adams DJ, Vaillancourt SM, Kaptoge S, Baldock P, Cooper C, Reeve J, Ntzani EE, Evangelou E, Ohlsson C, Karasik D, Rivadeneira F, Kiel DP, Tobias JH, Gregson CL, Harvey NC, Grundberg E, Goltzman D, Adams DJ, Lelliott CJ, Hinds DA, Ackert-Bicknell CL, Hsu YH, Maurano MT, Croucher PI, Williams GR, JHD B, Evans DM, Richards JB (2019) An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet 51(2):258–266. https://doi.org/10.1038/s41588-018-0302-x
Pei YF, Liu L, Liu TL, Yang XL, Zhang H, Wei XT, Feng GJ, Hai R, Ran S, Zhang L (2019) Joint association analysis identified 18 new loci for bone mineral density. J Bone Miner Res 34(6):1086–1094. https://doi.org/10.1002/jbmr.3681
Liu YZ, Pei YF, Liu JF, Yang F, Guo Y, Zhang L, Liu XG, Yan H, Wang L, Zhang YP, Levy S, Recker RR, Deng HW (2009) Powerful bivariate genome-wide association analyses suggest the SOX6 gene influencing both obesity and osteoporosis phenotypes in males. PLoS One 4(8):e6827. https://doi.org/10.1371/journal.pone.0006827
Tan LJ, Wang ZE, Wu KH, Chen XD, Zhu H, Lu S, Tian Q, Liu XG, Papasian CJ, Deng HW (2015) Bivariate genome-wide association study implicates ATP6V1G1 as a novel pleiotropic locus underlying osteoporosis and age at menarche. J Clin Endocrinol Metab 100(11):E1457–E1466. https://doi.org/10.1210/jc.2015-2095
Design of the Women's Health Initiative clinical trial and observational study. The Women's Health Initiative Study Group (1998) Control Clin Trials 19(1):61–109. https://doi.org/10.1016/s0197-2456(97)00078-0
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575. https://doi.org/10.1086/519795
Genomes Project C, Abecasis GR, Auton A, Brooks LD, MA DP, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491(7422):56–65. https://doi.org/10.1038/nature11632
Zhang L, Pei YF, Fu X, Lin Y, Wang YP, Deng HW (2014) FISH: fast and accurate diploid genotype imputation via segmental hidden Markov model. Bioinformatics 30(13):1876–1883. https://doi.org/10.1093/bioinformatics/btu143
Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34(8):816–834. https://doi.org/10.1002/gepi.20533
Zhang L, Pei YF, Li J, Papasian CJ, Deng HW (2009) Univariate/multivariate genome-wide association scans using data from families and unrelated samples. PLoS One 4(8):e6502. https://doi.org/10.1371/journal.pone.0006502
Konstantopoulos S (2006) Fixed and mixed effects models in meta-analysis. IZA Discussion Paper No. 2198. Available at SSRN: https://ssrn.com/abstract=919993
Styrkarsdottir U, Stefansson OA, Gunnarsdottir K, Thorleifsson G, Lund SH, Stefansdottir L, Juliusson K, Agustsdottir AB, Zink F, Halldorsson GH, Ivarsdottir EV, Benonisdottir S, Jonsson H, Gylfason A, Norland K, Trajanoska K, Boer CG, Southam L, Leung JCS, Tang NLS, Kwok TCY, Lee JSW, Ho SC, Byrjalsen I, Center JR, Lee SH, Koh JM, Lohmander LS, Ho-Pham LT, Nguyen TV, Eisman JA, Woo J, Leung PC, Loughlin J, Zeggini E, Christiansen C, Rivadeneira F, van Meurs J, Uitterlinden AG, Mogensen B, Jonsson H, Ingvarsson T, Sigurdsson G, Benediktsson R, Sulem P, Jonsdottir I, Masson G, Holm H, Norddahl GL, Thorsteinsdottir U, Gudbjartsson DF, Stefansson K (2019) GWAS of bone size yields twelve loci that also affect height, BMD, osteoarthritis or fractures. Nat Commun 10(1):2054. https://doi.org/10.1038/s41467-019-09860-0
Li X, Zhu X (2017) Cross-phenotype association analysis using summary statistics from GWAS. Methods Mol Biol 1666:455–467. https://doi.org/10.1007/978-1-4939-7274-6_22
Codoni V, Blum Y, Civelek M, Proust C, Franzen O, Cardiogenics C, CADGenomics ILC, Bjorkegren JL, Le Goff W, Cambien F, Lusis AJ, Tregouet DA (2016) Preservation analysis of macrophage gene coexpression between human and mouse identifies PARK2 as a genetically controlled master regulator of oxidative phosphorylation in humans. G3 (Bethesda) 6(10):3361–3371. https://doi.org/10.1534/g3.116.033894
Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, Dolan ME, Cox NJ (2010) SCAN: SNP and copy number annotation. Bioinformatics 26(2):259–262. https://doi.org/10.1093/bioinformatics/btp644
Mullin BH, Zhu K, Xu J, Brown SJ, Mullin S, Tickner J, Pavlos NJ, Dudbridge F, Walsh JP, Wilson SG (2018) Expression quantitative trait locus study of bone mineral density GWAS variants in human osteoclasts. J Bone Miner Res 33(6):1044–1051. https://doi.org/10.1002/jbmr.3412
Ward LD, Kellis M (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40(Database issue):D930–D934. https://doi.org/10.1093/nar/gkr917
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/nar/gky1131
Medina-Gomez C, Kemp JP, Trajanoska K, Luan J, Chesi A, Ahluwalia TS, Mook-Kanamori DO, Ham A, Hartwig FP, Evans DS, Joro R, Nedeljkovic I, Zheng HF, Zhu K, Atalay M, Liu CT, Nethander M, Broer L, Porleifsson G, Mullin BH, Handelman SK, Nalls MA, Jessen LE, Heppe DHM, Richards JB, Wang C, Chawes B, Schraut KE, Amin N, Wareham N, Karasik D, Van der Velde N, Ikram MA, Zemel BS, Zhou Y, Carlsson CJ, Liu Y, McGuigan FE, Boer CG, Bonnelykke K, Ralston SH, Robbins JA, Walsh JP, Zillikens MC, Langenberg C, Li-Gao R, Williams FMK, Harris TB, Akesson K, Jackson RD, Sigurdsson G, den Heijer M, van der Eerden BCJ, van de Peppel J, Spector TD, Pennell C, Horta BL, Felix JF, Zhao JH, Wilson SG, de Mutsert R, Bisgaard H, Styrkarsdottir U, Jaddoe VW, Orwoll E, Lakka TA, Scott R, Grant SFA, Lorentzon M, van Duijn CM, Wilson JF, Stefansson K, Psaty BM, Kiel DP, Ohlsson C, Ntzani E, van Wijnen AJ, Forgetta V, Ghanbari M, Logan JG, Williams GR, Bassett JHD, Croucher PI, Evangelou E, Uitterlinden AG, Ackert-Bicknell CL, Tobias JH, Evans DM, Rivadeneira F (2018) Life-course genome-wide association study meta-analysis of total body BMD and assessment of age-specific effects. Am J Hum Genet 102(1):88–102. https://doi.org/10.1016/j.ajhg.2017.12.005
Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, Zhernakova A, Zhernakova DV, Veldink JH, Van den Berg LH, Karjalainen J, Withoff S, Uitterlinden AG, Hofman A, Rivadeneira F, Hoen PAC, Reinmaa E, Fischer K, Nelis M, Milani L, Melzer D, Ferrucci L, Singleton AB, Hernandez DG, Nalls MA, Homuth G, Nauck M, Radke D, Volker U, Perola M, Salomaa V, Brody J, Suchy-Dicey A, Gharib SA, Enquobahrie DA, Lumley T, Montgomery GW, Makino S, Prokisch H, Herder C, Roden M, Grallert H, Meitinger T, Strauch K, Li Y, Jansen RC, Visscher PM, Knight JC, Psaty BM, Ripatti S, Teumer A, Frayling TM, Metspalu A, van Meurs JBJ, Franke L (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45(10):1238–1243. https://doi.org/10.1038/ng.2756
Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie SM, Fuchs H, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu CW, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Newbigging S, Nutter LM, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, Tamura M, Trainor AG, Tudose I, Wakana S, Warren J, Wendling O, West DB, Wong L, Yoshiki A, International Mouse Phenotyping C, Jackson L, Infrastructure Nationale Phenomin ICdlS, Charles River L, MRC H, Toronto Centre for P, Wellcome Trust Sanger I, Center RB, MacArthur DG, Tocchini-Valentini GP, Gao X, Flicek P, Bradley A, Skarnes WC, Justice MJ, Parkinson HE, Moore M, Wells S, Braun RE, Svenson KL, de Angelis MH, Herault Y, Mohun T, Mallon AM, Henkelman RM, Brown SD, Adams DJ, Lloyd KC, McKerlie C, Beaudet AL, Bucan M, Murray SA (2016) High-throughput discovery of novel developmental phenotypes. Nature 537(7621):508–514. https://doi.org/10.1038/nature19356
Chalmers TC, Smith H Jr, Blackburn B, Silverman B, Schroeder B, Reitman D, Ambroz A (1981) A method for assessing the quality of a randomized control trial. Control Clin Trials 2(1):31–49. https://doi.org/10.1016/0197-2456(81)90056-8
Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, Gagnon DR, DuVall SL, Li J, Peloso GM, Chaffin M, Small AM, Huang J, Tang H, Lynch JA, Ho YL, Liu DJ, Emdin CA, Li AH, Huffman JE, Lee JS, Natarajan P, Chowdhury R, Saleheen D, Vujkovic M, Baras A, Pyarajan S, Di Angelantonio E, Neale BM, Naheed A, Khera AV, Danesh J, Chang KM, Abecasis G, Willer C, Dewey FE, Carey DJ, Global Lipids Genetics C, Myocardial Infarction Genetics C, Geisinger-Regeneron Discov EHRC, Program VAMV, Concato J, Gaziano JM, O'Donnell CJ, Tsao PS, Kathiresan S, Rader DJ, Wilson PWF, Assimes TL (2018) Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet 50(11):1514–1523. https://doi.org/10.1038/s41588-018-0222-9
Liu K, Tan LJ, Wang P, Chen XD, Zhu LH, Zeng Q, Hu Y, Deng HW (2017) Functional relevance for associations between osteoporosis and genetic variants. PLoS One 12(4):e0174808. https://doi.org/10.1371/journal.pone.0174808
Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, Schoech A, Pasaniuc B, Price AL (2019) Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet 104(1):65–75. https://doi.org/10.1016/j.ajhg.2018.11.008
Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474(7351):337–342. https://doi.org/10.1038/nature10163
Tolksdorf F, Mikulec J, Geers B, Endig J, Sprezyna P, Heukamp LC, Knolle PA, Kolanus W, Diehl L (2018) The PDL1-inducible GTPase Arl4d controls T effector function by limiting IL-2 production. Sci Rep 8(1):16123. https://doi.org/10.1038/s41598-018-34522-4
Liu YZ, Dvornyk V, Lu Y, Shen H, Lappe JM, Recker RR, Deng HW (2005) A novel pathophysiological mechanism for osteoporosis suggested by an in vivo gene expression study of circulating monocytes. J Biol Chem 280(32):29011–29016. https://doi.org/10.1074/jbc.M501164200
Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K (2010) Development of monocytes, macrophages, and dendritic cells. Science 327(5966):656–661. https://doi.org/10.1126/science.1178331
Deng FY, Lei SF, Zhang Y, Zhang YL, Zheng YP, Zhang LS, Pan R, Wang L, Tian Q, Shen H, Zhao M, Lundberg YW, Liu YZ, Papasian CJ, Deng HW (2011) Peripheral blood monocyte-expressed ANXA2 gene is involved in pathogenesis of osteoporosis in humans. Mol cell proteomics 10(11):M111 011700. https://doi.org/10.1074/mcp.M111.011700
Manabe N, Kawaguchi H, Chikuda H, Miyaura C, Inada M, Nagai R, Nabeshima Y, Nakamura K, Sinclair AM, Scheuermann RH, Kuro-o M (2001) Connection between B lymphocyte and osteoclast differentiation pathways. J Immunol 167(5):2625–2631. https://doi.org/10.4049/jimmunol.167.5.2625
Jowsey J, Detenbeck LC (1969) Importance of thyroid hormones in bone metabolism and calcium homeostasis. Endocrinology 85(1):87–95. https://doi.org/10.1210/endo-85-1-87
Murphy E, Williams GR (2004) The thyroid and the skeleton. Clin Endocrinol 61(3):285–298. https://doi.org/10.1111/j.1365-2265.2004.02053.x
Acknowledgments
We appreciate all the volunteers who participated into this study. We are grateful to both GEFOS consortia and the deCODE study for releasing large-scale GWAS summary results for replication analysis.
Funding
LZ and YFP are partially supported by the National Natural Science Foundation of China (31571291, 31771417, and 31501026) and a project of the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions. RH is partially supported by the Inner Mongolia Autonomous Region Medical Health Science & Technology Research Program (201702180). HS and HWD are partially supported by the National Institutes of Health (R01AR059781, P20GM109036, R01MH107354, R01MH104680, R01GM109068, R01AR069055, U19AG055373, R01DK115679), the Edward G. Schlieder Endowment and the Drs. W. C. Tsai and P. T. Kung Professorship in Biostatistics from Tulane University. The numerical calculations in this paper have been done on the supercomputing system of the National Supercomputing Center in Changsha. The WHI program is funded by the National Heart, Lung, and Blood Institute, National 20 Institutes of Health, US Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. This manuscript was not prepared in collaboration with investigators of the WHI, has not been reviewed and/or approved by the Women’s Health Initiative (WHI), and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. Funding for WHI SHARe genotyping was provided by NHLBI Contract N02-HL-64278. The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000200.v10.p3.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
None
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 149 kb)
Rights and permissions
About this article
Cite this article
Zhang, H., Liu, L., Ni, JJ. et al. Pleiotropic loci underlying bone mineral density and bone size identified by a bivariate genome-wide association analysis. Osteoporos Int 31, 1691–1701 (2020). https://doi.org/10.1007/s00198-020-05389-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00198-020-05389-x