Abstract
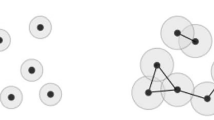
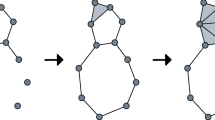
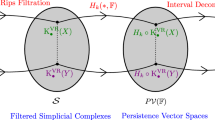
In this article, we define a new non-archimedean metric structure, called cophenetic metric, on persistent homology classes of all degrees. We then show that zeroth persistent homology together with the cophenetic metric and hierarchical clustering algorithms with a number of different metrics do deliver statistically verifiable commensurate topological information based on experimental results we obtained on different datasets. We also observe that the resulting clusters coming from cophenetic distance do shine in terms of different evaluation measures such as silhouette score and the Rand index. Moreover, since the cophenetic metric is defined for all homology degrees, one can now display the inter-relations of persistent homology classes in all degrees via rooted trees.







Similar content being viewed by others
Notes
The source code and the data of the numerical experiments we conducted in the paper can be found on the authors’ GitHub page at https://github.com/ismailguzel/TDA-HC .
The computational tools we use in this section are as follows: To compare dendrograms, we use tools come from the dendextend (Galili 2015) and vegan (Oksanen et al. 2019) packages of the R programming language (R Core Team 2021). For the map of Turkey, we used Generic Mapping Tools (Wessel et al. 2019). To compute cophenetic distance matrix we used SageMath Developers et al. (2020). In order to compute and visualize clusters, we used python programming language (Van Rossum and Drake 2009) and its scikit-learn library (Pedregosa et al. 2011).
References
Adams H, Emerson T, Kirby M, Neville R, Peterson C, Shipman P, Chepushtanova S, Hanson E, Motta F, Ziegelmeier L (2017) Persistence images: a stable vector representation of persistent homology. J Mach Learn Res 18(1):218–252
Agresti A (2019) An introduction to categorical data analysis. Wiley Series in Probability and Statistics. John Wiley & Sons, Inc., Hoboken, NJ. Third edition of [ MR1394195]
Ben-Hur A, Horn D, Siegelmann HT, Vapnik V (2002) Support vector clustering. J Mach Learn Res 2(2):125–137
Bray JR, Curtis JT (1957) An ordination of upland forest communities of southern wisconsin. Ecol Monogr 27:325–349
Bubenik P (2015) Statistical topological data analysis using persistence landscapes. J Mach Learni Res 16(1):77–102
Buchin K, Buchin M, Byrka J, Nöllenburg M, Okamoto Y, Silveira RI, Wolff A (2012) Drawing (complete) binary tanglegrams: hardness, approximation, fixed-parameter tractability. Algorithmica 62(1–2):309–332
Carlsson G (2020) Persistent homology and applied homotopy theory. In: Handbook of Homotopy Theory, pp. 297–329. Chapman and Hall/CRC
Carlsson G, Mémoli F (2008) Persistent clustering and a theorem of j. kleinberg. arXiv preprint arXiv:0808.2241
Carlsson G, Mémoli F (2010) Characterization, stability and convergence of hierarchical clustering methods. J Mach Learn Res 11:1425–1470
Carlsson G, Zomorodian A, Collins A, Guibas LJ (2005) Persistence barcodes for shapes. Int J Shape Model 11(02):149–187
Chung YM, Lawson A (2019) Persistence curves: A canonical framework for summarizing persistence diagrams. arXiv preprint arXiv:1904.07768
Cohen-Steiner D, Edelsbrunner H, Harer J (2007) Stability of persistence diagrams. Discret Comput Geom 37(1):103–120
Developers TS, Stein W, Joyner D, Kohel D, Cremona J, Eröcal B (2020) Sagemath, version 9.0
Donaldson S (2011) Riemann surfaces, Oxford Graduate Texts in Mathematics, vol 22. Oxford University Press, Oxford
Edelsbrunner H, Letscher D, Zomorodian A (2000) Topological persistence and simplification. In: Proceedings 41st annual symposium on foundations of computer science, pp. 454– 463. IEEE
Elkin Y, Kurlin V (2020) The mergegram of a dendrogram and its stability. arXiv preprint arXiv:2007.11278
Fernau H, Kaufmann M, Poths M (2010) Comparing trees via crossing minimization. J Comput Syst Sci 76(7):593–608
Fisher RA (1936) The use of multiple measurements in taxonomic problems. Ann Eugen 7(2):179–188
Galili T (2015) dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 31(22):3718–3720
Ghrist R (2008) Barcodes: the persistent topology of data. Am Math Soc, Bull, New Ser 45(1):61–75
Hartigan JA (1985) Statistical theory in clustering. J Classif 2(1):63–76
Hubert L, Arabie P (1985) Comparing partitions. J Classif 2(1):193–218
Ignacio PSP (2020) Intrinsic hierarchical clustering behavior recovers higher dimensional shape information. arXiv preprint arXiv:2010.03894
Jain AK, Dubes RC (1988) Algorithms for clustering data. Prentice Hall Advanced Reference Series. Prentice Hall Inc, Englewood Cliffs, NJ
Jardine N, Sibson R (1971) Mathematical taxonomy. John Wiley & Sons Ltd., London-New York-Sydney. Wiley Series in Probability and Mathematical Statistics
Johnson SC (1967) Hierarchical clustering schemes. Psychometrika 32(3):241–254
Kleinberg JM (2002) An impossibility theorem for clustering. In: Becker S, Thrun S, Obermayer K (eds) Advances in neural information processing systems 32. MIT Press, Neural Information Processing Systems, pp 446–453
Kuhn HW (2005) The hungarian method for the assignment problem. Naval Res Logist (NRL) 52(1):7–21
Lance GN, Williams WT (1967) A general theory of classificatory sorting strategies: 1. hierarchical systems. Comput J 9(4):373–380
Legendre P, Legendre L (2012) Numerical ecology, 3rd edn. Elsevier
Lumbreras A, Velcin J, Guégan M, Jouve B (2017) Non-parametric clustering over user features and latent behavioral functions with dual-view mixture models. Comput Stat 32(1):145–177
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Canc Res 27:209–220
Melnykov V, Zhu X (2019) An extension of the \(K\)-means algorithm to clustering skewed data. Comput Stat 34(1):373–394
Merelli E, Rucco M, Sloot P, Tesei L (2015) Topological characterization of complex systems: Using persistent entropy. Entropy 17(10):6872–6892
Miller H (2020) Handbook of homotopy theory. CRC Press/Chapman and Hall Handbooks in Mathematics Series. CRC Press, Boca Raton, FL
Moon C, Giansiracusa N, Lazar NA (2018) Persistence terrace for topological inference of point cloud data. J Comput Gr Stat 27(3):576–586
Oksanen J, Blanchet FG, Friendly M, Kindt, R, Legendre P, McGlinn D, Minchin PR, O’Hara R, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2019) vegan: Community ecology package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan
Patrício M, Pereira J, Crisóstomo J, Matafome P, Gomes M, Seiça R, Caramelo F (2018) Using resistin, glucose, age and bmi to predict the presence of breast cancer. BMC Cancer 18:1–8
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E (2011) Scikit-learn: machine learning in python. J Mach Learn Res 12:2825–2830
R Core Team: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2021)
Rosenberg A, Hirschberg J (2007) V-measure: A conditional entropy-based external cluster evaluation measure. In: Proceedings of the 2007 joint conference on empirical methods in natural language processing and computational natural language learning (EMNLP-CoNLL), pp. 410–420
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65
Scornavacca C, Zickmann F, Huson DH (2011) Tanglegrams for rooted phylogenetic trees and networks. Bioinformatics 27(13):i248–i256
Sergios T, Konstantinos K (2009) Pattern recognition, fourth, edition. Academic Press, Boston
Sneath, PH, Sokal RR et al (1973) Numerical taxonomy. the principles and practice of numerical classification. W.H. Freeman and Company San Franscisco
Sokal RR, Rohlf FJ (1962) The comparison of dendrograms by objective methods. Taxon 11(2):33–40
Stong RE (1968) Notes on cobordism theory. Mathematical notes. Princeton University Press, Princeton, N.J.; University of Tokyo Press, Tokyo
Strehl A, Ghosh J (2002) Cluster ensembles-a knowledge reuse framework for combining multiple partitions. J Mach Learn Res 3:583–617
Van Rossum G, Drake FL (2009) Python 3 Reference Manual. CreateSpace, Scotts Valley, CA
Wessel P, Luis JF, Uieda L, Scharroo R, Wobbe F, Smith WHF, Tian D (2019) The generic mapping tools version 6. Geochem, Geophys, Geosyst 20(11):5556–5564
Zomorodian A, Carlsson G (2005) Computing persistent homology. Discret Comput Geom 33(2):249–274
Acknowledgements
The first author was supported by Research Fund Project Number TDK-2020-42698 of the Istanbul Technical University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Güzel, İ., Kaygun, A. A new non-archimedean metric on persistent homology. Comput Stat 37, 1963–1983 (2022). https://doi.org/10.1007/s00180-021-01187-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00180-021-01187-z