Abstract
Behavioral researchers have shown growing interest in structural equation model trees (SEM Trees), a new recursive partitioning-based technique for detecting population heterogeneity. In the present research, we conducted a large-scale simulation to investigate the performance of latent growth curve model (LGCM)-based SEM Trees for uncovering between-individual differences in patterns of within-individual change. Simulation results showed that the correct estimation rates of the number of classes are most strongly related to the agreement rate of the covariate with its true latent profile, and the number of true classes also has a serious negative impact on correct estimation rates of the number of classes. SEM Trees is not always sensitive to the influence of model misspecification, and its impact differs according to a complex function of the types of misspecification as well as the statistical properties of the template model. On the whole, LGCM-based SEM Trees is a robust and stable approach under possible model misspecifications.
Similar content being viewed by others
Behavioral researchers are often interested in investigating population heterogeneity (i.e., between-individual differences in within-individual change patterns) that may appear in longitudinal data. The primary research purpose is to understand the potential variety of typical development and growth trajectories. Ignoring population heterogeneity may lead to incorrect conclusions concerning development and growth trajectories. A large amount of research has revealed evidence of the presence of population heterogeneity in longitudinal trajectories. For example, Nagin and Tremblay (1999) extracted four classes representing different patterns in changes of boys’ physical aggression scores during childhood: a “low” class comprising children who display little or no physically aggressive behavior; a “moderate declining” class that displayed a modest level of physical aggression at age 6, but by age 10 had largely desisted; a “high declining” class that showed serious physical aggression at age 6 but scores far lower by age 15; and a “chronics” class comprised of children who displayed high levels of physical aggression throughout the observation period. Another example includes the work of Leiby et al. (2009), who detected three longitudinal patterns in perceived pain scores reported by patients with interstitial cystitis: a “responder” class that reported regular symptoms throughout the observation period, a “non-responder” class that showed large scores in the first weeks but far lower scores later in the observation period, and “temporary responders” who at first reported reduced symptoms but showed increased scores later (i.e., U-shaped profiles).
To uncover the potential population heterogeneity in longitudinal designs, various statistical techniques have been proposed, including non-hierarchical cluster analysis (Genolini and Falissard 2010; Usami 2014a), finite mixture models (McLachlan and Peel 2000; Todo and Usami 2016), the latent class model (e.g., Nagin 1999; Nagin and Land 1993; Nagin and Tremblay 1999, for a group-based semi-parametric approach), multi-group analysis (Little et al. 2000; McArdle and Nesselroade 2014), mixed-effects models (moderation; Preacher et al. 2016), and decision trees (Brandmaier et al. 2013, 2014; Usami et al. 2017, for structural equation model (SEM) trees, Sela and Simonoff 2012, for random effects (RE-EM) Trees, and Fokkema et al. (2017), for generalized linear mixed-effects model tree). During the past decade, researchers have shown growing interest in applying latent growth curve mixture models (LGCMMs; Berlin et al. 2014; Leiby et al. 2009; Neelon et al. 2011; Ram and Grimm 2009), and, more recently, in applying machine learning techniques including SEM Trees (e.g., Hayes et al. 2015; Martin 2015; Jacobucci et al. 2017). Both LGCMMs and SEM Trees utilize SEM to model changes using latent variables estimated with a smaller number of parameters.
LGCMMs combine (unstructured) finite mixture models (McLachlan and Peel 2000) and latent growth curve models (LGCM; Bollen and Curran 2006; Meredith and Tisak 1984, 1990), allowing researchers to investigate heterogeneity in longitudinal trajectories using a categorical latent variable called a class (also called a component, cluster, group, or regime). SEM Trees also synthesizes aspects of two statistical traditions, combining decision/regression trees (Morgan and Sonquist 1963; Sonquist and Morgan 1964) and SEM. SEM Trees explore population heterogeneity through partitioning the dataset based on the specific value of the observed covariate that results in the largest differences in parameter estimates returned by the prescribed SEM (called the template model or the hypothesized model; Brandmaier et al. 2013). SEM Trees is a supervised classification model that uses observed covariates for classification, whereas an LGCMM is an unsupervised classification model for which such observed covariates are not required. Because of this property, SEM Trees can find covariates and covariate interactions that predict nonlinear differences in structural parameters among the observed variables (Brandmaier et al. 2013).
SEM Trees continues splitting the data using covariate information through recursive partitioning, and can detect population heterogeneity that may appear in longitudinal trajectories characterized by a prespecified template model. The availability of useful covariates to explain population heterogeneity is thus key, since without such covariates the dataset cannot be partitioned, even if population heterogeneity exists. SEM Trees can be effectively applied as a data-mining tool in order to detect population heterogeneity when very large numbers of covariates are available (Brandmaier et al. 2014). As we will explain in the next section, the SEM Tree algorithm adds splits to a tree on the basis of likelihood-ratio tests. Note that the SEM Tree algorithm’s use of maximum likelihood in the discrepancy function to grow trees differs from the partitioning methods proposed by Sanchez (2009), which use (partial) least-squares estimation. SEM Trees analyses can be performed using the R package semtree (Brandmaier et al. 2013), with SEM models handled by either lavaan (Rosseel 2012) or OpenMx (Boker et al. 2011). In this package, we can also use a more advanced version of SEM Trees called SEM forests (Brandmaier et al. 2016), which are ensembles of SEM Trees based on resamplings of the original dataset that provide increased stability of the estimation results. Currently, SEM forests can only be paired with OpenMx.
Compared with LGCMMs, SEM Trees is a recent development that has received much less application and consideration. However, many researchers have shown increasing interest in both theoretical and applied domains (e.g., Hayes et al. 2015; Hayes and McArdle 2017; Martin 2015; Miller et al. 2015; Usami et al. 2017). In applying SEM Trees (and LGCMMs), a difficult but intriguing question is the estimation of the total number of classes in the latent space that explain the population heterogeneity. In SEM Trees, the number of classes is equal to the total number of child nodes in the estimated tree. Although various methods of constructing decision trees have been developed for multivariate data (e.g., the method proposed by Brodley and Utgoff 1995, which does not assume structural equations among variables), in SEM Trees the observed precision in detecting population heterogeneity may differ from other methods due to differences in the number of parameters and sensitivity to possible misspecification, which is more typical in SEM. Therefore, unfortunately little is known about the performance of SEM Trees (and other related methods).Footnote 1 On this point, Usami et al. (2017) used a (bivariate) latent-change score (LCS) model as a data generation model and investigated the conditions under which SEM Trees might better perform at correctly identifying the true classes (nodes). They found that (1) correct estimation rates of the number of classes were strongly related to the agreement rate of the covariate with its true latent profile, (2) the influence of total sample size were also notable, (3) influences of other manipulated factors including mixture proportions and degree of separation (distances) of intercept factors between classes were almost ignorable, and (4) trees were very sensitive to the influence of model misspecification with respect to the template model (e.g., correct estimation rates of the number of classes were zero under the influences of model misspecification in time-variant error variances and random intercepts).
Although the investigation of Usami et al. (2017) is useful to improve our understanding about the performance of estimation by SEM Trees, this simulation may unfortunately be rather limited in that only the (bivariate) LCS model was used as a data generation model. The (bivariate) LCS model adds autoregressive and coupling (cross-lagged) terms into LGCM, and it can be used for inferring longitudinal relationship between variables (see a later section regarding this point). Despite the generality of its model structure, the LCS model has unfortunately not been widely used so far, and LGCM has still gained growing attention from applied researchers who aim to model longitudinal trajectories. An additional negative aspect of the LCS model comes from its high frequency of estimation problems (i.e., non-convergence problems and improper solutions) and potentially seriously unstable estimates of autoregressive and coupling (cross-lagged) parameters (e.g., Usami et al. 2015), and this might explain the serious sensitivity of (bivariate) LCS model-based SEM Trees to the influence of model misspecification. For that reason, the LCS model-based SEM Trees may not necessarily be a reasonable choice in practice, and substantially different performance (e.g., the frequency of estimation problems, correct estimation rates of the number of classes, sensitivity to the influence of model misspecification) may be observed when applying LGCM-based SEM Trees due to the difference in statistical properties from the LCS model. Actually, in the context of finite mixture models and LGCMM, Todo and Usami (2016) showed that correct estimation rates were largely different according to the analysis models specified. Because model misspecification should typically arise in wide applications of LGCM (SEM) in a strict sense (on this point, see Bauer and Curran 2004), seeking possible (tree-based) alternatives from LCS model-based SEM Trees must be pragmatically important for researchers who aim to detect population heterogeneity in longitudinal trajectories based on covariates.
Furthermore, due to the computational burden of the simulation,Footnote 2 the number of time points and true classes (nodes) were not manipulated in Usami et al. (2017). However, only allowing for two classes, thus only one split without covariate interactions, is a very limited case in general applications of tree analyses. Actually, previous research that applied LGCMM or SEM Trees extracted more than two classes (e.g., see the above examples of Jacobucci et al. 2017; Leiby et al. 2009; Nagin and Tremblay 1999), and we expect that researchers usually collect multiple covariates that can explain population heterogeneity in practice. Importantly, as the number of true classes and covariates become larger, exact estimation of population heterogeneity should become more difficult, since all covariates and corresponding criterion values must be identified correctly, demanding larger sample sizes in total when estimating trees. Likewise, the number of time points in the dataset can also vary in actual applications, and such differences might relate to the stability of parameters in SEM. Therefore, the previous investigations are limited in addressing these points, and there might be great risk of drawing wrong conclusions about the performance of SEM Trees in general (e.g., correct estimation rates of the number of classes on average, or sensitivity to the influence of possible model misspecifications).
In this paper, our aim is to investigate the estimation performance of SEM Trees to correctly identify the true classes using linear and quadratic LGCM as a template model, manipulating the number of time points and classes (nodes) as well as the degree of separation, sample sizes, and agreement rates of the covariate with its true latent profile. We also consider the influence of model misspecification. These include wrong assumptions of the functional form of the development trajectories and time-invariant error variances. In the next section, we briefly explain the (linear and quadratic) LGCM and SEM Trees algorithm. We then present the simulation design and results, and close with suggested directions for future investigations.
1 LGCM and SEM Trees
1.1 LGCM
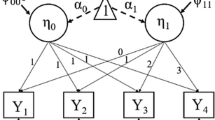
Let \(y_{it}\) be the observed variable at occasion t (\(1,\dots ,T\)) for each participant i (\(1,\dots ,N\)), and the quadratic LGCM expresses \(y_{it}\) as simple sum of true values as a function of time (\(f_{it}\)) and error (\(e_{it}\)). The quadratic LGCM can be written using a three-factor model as
where \(f_{it}=a_{0t}I_{i}+a_{1t}S_{1i}+a_{2t}S_{2i}\). \(a_{0t}\), \(a_{1t}\) and \(a_{2t}\) in the second line of the equation are factor loadings as functions of time, prespecified as \(a_{0t}=1\), \(a_{1t}=(t-1)\) and \(a_{2t}=(t-1)^2\), respectively. Substituting these values into the equation leads to the third line of the equation. \(I_{i}\), \(S_{1i}\) and \(S_{2i}\) are factor scores that characterize the initial value and amount of first-order and second-order changes of true latent trajectory of participant i, respectively. More specifically, considering \(f_{i1}=I_{i}\) at \(t=1\), \(I_i\) can be considered as the true value of participant i at \(t=1\). Interpretation of \(S_{1i}\) and \(S_{2i}\) can be better clarified by evaluating first- and second-order derivatives of \(f_{it}\) as \(f'_{it}=\partial f_{it}/\partial t=S_{1i}+2(t-1)S_{2i}\) and \(f''_{it}=\partial ^2 f_{it}/\partial t^2=2S_{2i}\). Considering that \(f'_{i1}=S_{1i}\) and \(f''_{it}\) is not a function of time (t), \(S_{1i}\) and \(S_{2i}\) can be more clearly interpreted as the rate of change (or “instant” slope) at \(t=1\) and (half of) its constant change per unit change of time. Thus, if \(S_{2i}=0\) for all participants, there is no change in the rate of change of the trajectory, so this defines a linear function (Bollen and Curran 2006).
Typically, an error term \(e_{it}\) is assumed to be normally distributed as \(N(0, \psi _t^2)\), and is also assumed to have no correlation with factor scores and errors of different participants \(i'\) at different time points \(t'\) (i.e., \(cor(I_i, e_{it})=cor(S_{1i}, e_{it})=cor(S_{2i}, e_{it})=0\), and \(cor(e_{it}, e_{i't})=cor(e_{it}, e_{it'}) =cor(e_{it}, e_{i't'})=0)\). Let \({\varvec{v}}\) and \({\varvec{\Phi }}\) be a factor means vector and variance–covariance matrix of latent factor scores \({\varvec{\xi _i}}=(I_i, S_{1i}, S_{2i})^t\), expressed as
When we aim to specify a linear LGCM by setting \(S_{2i}=0\) for all participants, this manipulation essentially indicates assumptions of zero means and zero (co)variances regarding the second-order slope factor (i.e., \(\mu _{S2}=\phi ^{2}_{S2}=\phi _{IS2}=\phi _{S1S2}=0\)).
Let \({\varvec{y}}\) be an observed data vector, and its elements be arranged as \({\varvec{y}}=({\varvec{y^t_{1}}},\dots ,{\varvec{y^t_{i}}},\dots ,{\varvec{y^t_{N}}})^t\), where \({\varvec{y_{i}}}=(y_{i1},\dots ,y_{it},\dots ,y_{iT})^t\). Then the likelihood function for \({\varvec{y}}\), which will be used in SEM Trees for maximum likelihood (ML) estimation, can be expressed as
where \({\varvec{\mu (\Theta )}}\) and \({\varvec{\Sigma (\Theta )}}\) are mean and covariance structures implied by the LGCM, respectively, and \({\varvec{\Theta }}\) represents all parameters included in the LGCM. The explicit link between \({\varvec{\mu (\Theta )}}\), \({\varvec{\Sigma (\Theta )}}\), and the parameters of the quadratic LGCM is provided in the literature (e.g., Bollen and Curran 2006, p. 92–93).
The (univariate) LCS model that assumes second-order changes (\(S_2\)) can be expressed as
where \(\beta ^*=1+\beta \), and \(\beta \) represents an autoregressive parameter. Factor loadings \(a^*_{1t}\) and \(a^*_{2t}\) are prespecified as \(a^*_{1t}=1\) and \(a^*_{2t}=(t-1)\), respectively. McArdle (2009) and Usami et al. (2015) noted that the LGCM model can be considered as a special case of the LCS model. Actually, specifying \(\beta =0\) in Eq. (4) leads to the expression \(y_{it}=I_{i}+(t-1)S_{1i}+(t-1)^2S_{2i}+e_{it}\), which is identical to Eq. (1). In the bivariate case, although the coupling (cross-lagged) parameter is typically included in the LCS that reflects a longitudinal relationship between variables, the nested relationship between the LCS model and the LCGM can be derived in the same manner. With an empirical data example, Usami et al. (2015) argued that the LCS model might cause a high frequency of estimation problems (i.e., non-convergence problems and improper solutions) and potentially seriously unstable estimates of autoregressive and coupling (cross-lagged) parameters. A more detailed explanation of these models that uses path diagrams is provided in Usami et al. (2015).
1.2 SEM Trees
The estimation procedure in SEM Trees can be summarized as follows (Brandmaier et al. 2013; Usami et al. 2017):
-
(i)
Fit an SEM (template model) to the whole dataset and calculate the likelihood \(L({\varvec{\Theta }}|{\varvec{y}})\).
-
(ii)
Search for a covariate M and corresponding criterion value m that maximizes the heterogeneity of the SEM parameters in a round-robin fashion. Specifically, such M and m maximize the following likelihood \(L^*({\varvec{\Theta _A,\Theta _B}}|{\varvec{y}})\):
$$\begin{aligned} L^*({\varvec{\Theta _A,\Theta _B}}|{\varvec{y}})=L({\varvec{\Theta _A}}|{\varvec{y_A}}, M\ge m)+L({\varvec{\Theta _B}}|{\varvec{y_B}}, M< m), \end{aligned}$$(5)by splitting the whole dataset into subgroups A and B. Here, \(L({\varvec{\Theta _A}}|{\varvec{y_A}}, M\ge m)\) is a likelihood for subgroup A with observed longitudinal data \(y_A\), which satisfies the condition \(M\ge m\), and \(L({\varvec{\Theta _B}}|{\varvec{y_B}}, M< m)\) is a likelihood for subgroup B with observed longitudinal data \(y_B\), which satisfies the condition \(M<m\), respectively. \({\varvec{\Theta _A}}\) and \({\varvec{\Theta _B}}\) are SEM parameters corresponding to subgroups A and B, respectively.
-
(iii)
Investigate whether the model fits significantly better by splitting the data. Specifically, the observed difference in (log-)likelihood
$$\begin{aligned} \chi ^2=2\log L^*({\varvec{\Theta _A,\Theta _B}}|{\varvec{y}})-2\log L({\varvec{\Theta }}|{\varvec{y}}), \end{aligned}$$(6)is evaluated using a chi-square distribution with degrees of freedom equal to the number added by having two models versus one under the prespecified significance level.
-
(iv)
The whole dataset is split into subgroups A and B if \(\chi ^2\) exceeds the corresponding critical value. If not, the dataset is not split and estimation terminates. In the former case, repeat steps (i)–(iii) for both subgroups A and B and split the data if \(\chi ^2\) significantly improves. Repeat this recursive partitioning calculation process and continue splitting the data until \(\chi ^2\) does not show statistical improvement.
-
(v)
To solve the problem of multiple comparisons and type-I error inflation resulting from over-splitting the data, either a Bonferroni correction or cross-validation (CV) procedure can be used to remove unnecessary subgroups (child nodes) if they exist. Then, estimation is finished and the total number of remaining child nodes becomes an estimate of the number of classes in SEM Trees.
Due to the procedure in (ii) [i.e., Eq. (5)], the algorithm of SEM Trees is typically computationally intensive since a new model has to be estimated for every possible criterion value. Note that nominal data (e.g., race categories) \(M^*\) can also be included as covariates in SEM Trees (e.g., Brandmaier et al. 2013). In that case, data is split on the basis of whether a participant belongs to a specific set A (e.g., Asian or American Indian) or its complement set B (=\({\bar{A}}\)), and a likelihood similar to Eq. (5) is constructed as \(L^*({\varvec{\Theta _A,\Theta _B}}|{\varvec{y}})=L({\varvec{\Theta _A}}|{\varvec{y_A}}, M^*\in A)+L({\varvec{\Theta _B}}|{\varvec{y_B}}, M^* \in B)\).
2 Simulation
In this simulation, we investigate the estimation performance of SEM Trees to correctly identify the true classes using linear and quadratic LGCMs under the influence of model misspecification of the functional form of the development trajectories and time-invariant error variances. We also conducted several supplemental simulations to confirm the generalizability of the main simulation and to briefly compare the performance of LGCMMs with SEM Trees. Simulation code and result sheets are available in the supplemental online materials.
2.1 Manipulated factors
For data generation, we systematically changed the number of total participants (\(N= 100, 200, 400, 800, 1600, 3200\)), the number of time points (\(T= 4, 6, 8\)), the number of true classes (\(C= 2, 4\)), the degree of separation (i.e., distances) between classes at the first time-point (\(d= 0.5, 1.0, 1.5\), 2.0, 2.5), the agreement rate of the observed dichotomous covariate with its true latent profile (\(r= 0.5, 0.6, 0.7, 0.8, 0.9, 1.0\); an index that indicates the extent of association between covariate and class differences—see the explanation below for more details), and a functional form of true latent trajectory in each class (linear or quadratic curve). We specified the levels of these factors to range from small to large values so that the simulation would cover various kinds of developmental trajectories that appear in actual longitudinal data. Additionally, we manipulated the presence or absence of two kinds of model misspecification: incorrect functional forms (e.g., a model that incorrectly assumes a linear latent trajectory is fit to a dataset that actually has a quadratic functional form, or vice versa) and time-invariant error variances (i.e., a model that assumes time-invariant error variances is incorrectly fit to a dataset that was actually generated using time-variant error variances). We generated simulation data under an orthogonal design. Namely, there are (\(6 \times 3 \times 2 \times 5 \times 6 \times 2 \times 2=\)) 4320 distinct combinations of factors. Additionally, 100 sets of simulation data were randomly generated for each condition, yielding \(4320 \times 100=432{,}000\) datasets.
We assumed researchers have either one or two dichotomous covariate(s) in the dataset when \(C=2\) and \(C=4\), respectively. Using dichotomous covariates is advantageous in this simulation, in that they can simplify the procedure of evaluating the performance of estimating the number of classes when \(C=4\), because the correct estimation of the number of classes (\({\hat{C}}=4\)) indicates the correct extraction of the true classification structure (i.e., data splits into two nodes by a covariate, and each node is further split by another covariate, resulting in four classes). Therefore, in this simulation setting, incorrect estimation of the number of classes indicates underestimation of classes.
In addition, we did not manipulate the proportion of class sizes (e.g., a 50–50% allocation of participants in two classes, or a 90–10% allocation) and proportions were assumed to be equal among classes, because previous research (e.g., Henson et al. 2007: Usami 2014b) that investigated the performance of LGCMMs has shown that differences in the proportion of class sizes are not influential.
2.2 Data generation
Simulation data were generated through the following procedure using actual parameter estimates from child weight data of \(T=5\) in the National Longitudinal Survey of Youth (Biesanz et al. 2004; cf Bollen and Curran 2006, pp. 94–96). (i) Under a fixed number of true classes (C) and time points (T), we set slope factor means to \(\mu _{I}=39.563\), \(\mu _{S1}=6.988\), and \(\mu _{S2}=0.373\). Class-invariant variance–covariance matrix \({\varvec{\Phi }}\) was specified as
Note that when the functional form of true latent trajectory is linear, zero factor means and zero (co)variances are assumed (i.e., \(\mu _{S2}=\phi ^{2}_{S2}=\phi _{IS2}=\phi _{S1S2}=0\)). (Time-variant) error variances \(\psi _t^2\) are specified as \(\psi _1^2=2.942, \psi _2^2=15.084, \psi _3^2=44.858, \psi _4^2=85.200, \psi _5^2=73.285, \psi _6^2=89.280, \psi _7^2=91.947\), and \(\psi _8^2=89.336\), irrespective of the conditions of T (e.g., when \(T=4\), the first four values \(\psi _1^2=2.942\), \(\psi _2^2=15.084\), \(\psi _3^2=44.858\), \(\psi _4^2=85.200\) are used).Footnote 3 (ii) Let d be a symbol to adjust the expected degree of separation (i.e., the standardized mean differences) at \(t=1\) among classes, and set the intercept factor means as \(\mu ^{1}_{I}=\mu _{I}-0.5\times d\times \sigma _1\) and \(\mu ^{2}_{I}=\mu _{I}+0.5\times d\times \sigma _1\) in \(C=2\), and \(\mu ^{1}_{I}=\mu _{I}-1.5\times d\times \sigma _1\), \(\mu ^{2}_{I}=\mu _{I}-0.5\times d\times \sigma _1\), \(\mu ^{3}_{I}=\mu _{I}+0.5\times d\times \sigma _1\) and \(\mu ^{4}_{I}=\mu _{I}+1.5\times d\times \sigma _1\) in \(C=4\). Here, \(\sigma _1\) denotes (class-invariant) population standard deviations of the variables at \(t=1\), which can be calculated as \(\sigma _{1}=\sqrt{\phi _I^2+\psi _1^2}=\sqrt{33.913+2.942}=6.071\). Slope factor means (\(\mu _{S1}\), \(\mu _{S2}\)) are assumed to be class invariant, indicating true latent trajectories have the same degree of separations among classes in each time point. (iii) Using model parameters specified in the above steps, mean and variance–covariance structures (\({\varvec{\mu ^{c}(\Theta )}}\), \({\varvec{\Sigma ^{c}(\Theta )}}\)) are calculated for each class. (iv) Under the specified number of total participants N, generate data \({\varvec{y}}\) with size N / C using a multivariate normal distribution as \({\varvec{y^{c}_i}}\sim MVN({\varvec{\mu ^{c}(\Theta )}},{\varvec{\Sigma ^{c}(\Theta )}})\) for each class. (v) For the analysis of SEM Trees, generate dummy (discrete) covariates (each valued 0–1), which shows the \(100\times r\) % expected agreement rate for a true latent profile. Thus, when \(r=1\) the covariate essentially indicates a true latent profile for each participant, while \(r=0.50\) corresponds to the chance level (i.e., the covariate shows no correlation with the true latent profile). Thus, the larger r becomes, the more informative the covariate becomes in explaining population heterogeneity in the data.Footnote 4 When class sizes are equal, conditions of \(r=0.5, 0.6, 0.7, 0.8, 0.9, 1\) respectively correspond to expected correlations of 0, 0.20, 0.40, 0.60, 0.80 and 1 between true latent profiles (coded as 0 or 1 for two classes split from each node) and the dichotomous observed covariates. Complete datasets that include both observed variables \({\varvec{y}}\) and covariates are then generated.
2.3 Data analysis procedure
When fitting SEM Trees, we used three types of models in this simulation. The first is the linear or quadratic LGCM described by Eqs. (1)–(2) (i.e., the correctly specified model). The second is the LGCM incorrectly assuming time-invariant error variances (i.e., the wrong assumption that \(\psi _t^2=\psi ^2\)). The third is mathematically the same as the first, although the functional form is incorrectly specified (i.e., a model that assumes a linear latent trajectory is incorrectly fit to a dataset that actually has a quadratic functional form, or vice versa). The whole simulation procedure was performed in the R statistical environment (R Core Team 2016), and parameter estimation was conducted using the semtree package (Brandmaier et al. 2013) handled by lavaan (Rosseel 2012). In semtree, we used the naive method (i.e., splitting criterion based on the likelihood ratio test) and chose the Bonferroni correction for multiple testing in estimating trees.Footnote 5
2.4 Model selection procedure
In SEM Trees, the number of classes was estimated via the number of child nodes obtained without specifying a minimum number of within-node observations when growing the trees. The significance level was specified as 0.05. Estimation of the number of classes was considered correct if the dataset was correctly partitioned by a single covariate for \(C=2\) or by two covariates for \(C=4\), respectively.
2.5 Result
Table 1 shows the correct marginal estimation rates in each level of the manipulated factors according to the functional forms of the true latent trajectories (i.e., linear or quadratic) in the data generation model. Improper solutions were observed in 5–8% of cases when fitting quadratic LGCM assuming correct error variances (i.e., time-variant error variances) in \(T=4\), and repetitions were added until the total repetitions reached 100 in each condition. Irrespective of function form, the number of time points, which was newly manipulated in the current simulation, was almost unrelated to class-detection performance, indicating changes in this factor were not influential to statistical power (expected chi-square difference) when splitting nodes. However, the influences of other factors that characterize the properties of longitudinal data were notable. For example, larger values of the number of true nodes C, which is also a newly specified factor in the current simulation, have a negative impact on correct estimation of the number of true classes, because when \(C=4\), statistical power for correct estimation of classes dramatically decreases compared with the case of \(C=2\). This point is critically important, since in many applications of tree analyses there is more than one split, extracting more than two classes (e.g., Brandmaier et al. 2013; Jacobucci et al. 2017). Therefore, the previous investigations might show overly optimistic results regarding the performance of SEM Trees in general.
The agreement rate between the observed covariate and latent profile r also showed a dominant influence on the correct estimation rates, and values of r larger than 0.8 or 0.9 can be considered a minimum requirement for correctly detecting classes in the present context. Specifically, when the proportion of class sizes is equal (e.g., a 50–50% allocation of participants in classes), conditions of \(r=0.8\) and \(r=0.9\) correspond to expected correlations of 0.60 and 0.80 between true latent profiles and covariates, indicating that covariates that are substantially informative for predicting the true latent profile are required in applying SEM Trees. The total sample size N and degree of separation d were also influential on correct estimation rates, and \(d=1.5\) was roughly enough to maximize the performance of SEM Trees, given the other factors considered here. These results are almost identical, irrespective of the functional forms of the true latent trajectories. Note that the degree of separation was almost unrelated to the performance of SEM Trees in Usami et al. (2017), which used LCS model-based SEM Trees and manipulated differences in the intercept factor means. This might be attributed to unstable parameter estimates and the larger risk of improper solutions (e.g., local maxima) in the LCS model and the non-parallel and non-equidistant features of the true latent trajectories among the classes, resulting from parameters that are all class-variant specified in that research.
Irrespective of the functional form of true latent trajectories, influence of misspecification of the functional form was almost negligible. This can be partly attributed to what both linear and quadratic LGCMs share in common. Namely, both linear and quadratic LGCMs can express the monotone increasing trajectories that the simulated longitudinal dataset indicate. Interestingly, from Table 1 misspecification of the time-invariant error variances had larger impacts on the performance of SEM Trees, showing on average 6 and 8% decrease rates in linear and quadratic true latent trajectories, respectively. The misspecification exerted an influence on the estimated parameters, degrading model fit for each participant and ultimately causing problems when SEM Trees attempted to partition the dataset in search of a better-fitting tree. Statistical power in identifying classes decreased, resulting from smaller differences in parameters prescribed by the template model between classes due to model misspecification. However, Usami et al. (2017) showed much more serious impacts of model misspecification of error variances on correct estimation rates (e.g., correct estimation rates of the number of classes were zero), and this difference can be attributed to the frequent improper solutions and inflated standard errors of the autoregressive and cross-lagged parameters in LCS models, as well as the non-equidistant and non-parallel properties of the trajectories among classes in that research.
To facilitate a simple quantitative interpretation of how much each factor explained performance, we set the estimation results (i.e., rates of correct class identification) as a dependent variable and performed ANOVA, according to the functional form of true latent trajectory (linear or quadratic). A summary of the results is provided in Table 2. Note that the results for second-order to sixth-order interactions are omitted to save space. This table clearly shows the relatively dominant influence of the agreement rate between the observed covariate and latent profile r (\(SS=447.1, MS=89.41\), and \(\eta ^2=0.508\)), followed by the sample sizes N (\(SS=111.0, MS=22.19\), and \(\eta ^2=0.126\)) and the number of true classes C (\(SS=64.6, MS=64.64\), and \(\eta ^2=0.073\)). As we have seen in Table 1, the influence of C is dominant and it shows the second largest mean squares. However, influences of the two kinds of model misspecification are relatively small, and the number of time points T also showed small influence. These results indicate that in detecting population heterogeneity in the latent trajectories identified by SEM Trees, it is essential to collect covariates that are informative to predict true classification structures (i.e., larger agreement rates) and to collect sufficiently large sample sizes. Additionally, SEM Trees was not always sensitive to the influence of model misspecification, and its impacts were different according to a complex function of the type of misspecification and the statistical properties of the template model (stability of parameters and frequency of estimation problems in general).
In addition to the main effects, several first-order interactions relevant to agreement rates r (i.e., \(r\times C, r\times N, r\times d\)) are also notable. Regarding this point, Fig. 1 shows shifts in the correct marginal estimation rates at each level of the agreement rates under different true numbers of classes (C), sample sizes (N) and degrees of separations (d). From this figure, to achieve 0.80 or more correct estimation rates in the current simulation design, covariates that satisfy \(r=0.8\) or more are required when \(C=2\), whereas required r becomes more severe as \(r=0.95\) when \(C=4\), again indicating the difficulty of precise estimation of trees when C becomes larger. Likewise, to achieve very high correct estimation rates of 0.80 or more, at least \(r=0.9\) is required if the sample size is small (\(N=200\), \(N=400\)). However, required r becomes smaller as \(r=0.8\) and \(r=0.7\) if N becomes greater as \(N=800\) and \(N=1600\), respectively. When useful covariates are not gathered and r is small as \(r=0.6\), achieving 0.80 or more average correct estimation rates seems to be very difficult, even in extreme cases as simpler classification structures (i.e., \(C=2\)), large sample size (e.g., \(N=3200\)), and large degrees of separation (i.e., \(d=2.5\)).
2.6 Supplemental simulations
Further simulations are conducted under \(C=1\) (i.e., no population heterogeneity in true latent trajectory) with single dichotomous covariate and all other conditions being equal to investigate how frequently SEM Trees might overestimate the number of classes.Footnote 6 As a result, average correct estimation rates were 97%, and in all conditions SEM Trees could correctly estimate the number of classes in more than 95% of cases (see Table 1 in the supplemental online materials for details). Although this result indicates a low risk of SEM Trees overestimating the number of classes, the correct estimation rates in \(C=1\) should be decreased if the number of covariates increases due to an inflated type-1 error. On this point, if K covariates are mutually independent, from this result the rates can be conventionally estimated by \(0.97^K\). Thus, when the numbers of covariates are \(K=5\) and \(K=10\), the rates decrease to 86 and 74% on average, respectively, under the same procedure (i.e., Bonferroni correction). Note that in \(C=1\), misspecification of residual variances positively influences correct estimation rates, because in this case statistical power decreases for splitting the nodes.
We also assumed the presence of between-class differences in slope, and performed a similar simulation, limited to conditions of \(r=0.5,0.7,0.9\), \(N=100,400,1600\) and \(d=0.5,1.5,2.5\).Footnote 7 Correct marginal estimation rates are shown in Tables 2 and 3 of the supplemental online materials for the condition of intermediate and large between-class differences in slope means, respectively. Comparing the condition of no between-class differences in slope means (Table 4 of the supplemental online materials, which was created by removing the results of conditions \(r=0.6,0.8,1.0\), \(N=200,800,3200\), and \(d=1.0,2.0\) in the main simulation ), Tables 2 and 3 of the supplemental online materials show slightly higher correct estimation rates on average, but also show similar tendencies. Because it is natural to assume nonzero between-class differences in slope means (i.e., true latent trajectories in classes are not exactly parallel), correct estimation rates in Fig. 1 might indicate conservative values of rates, and rates might actually take larger values. A similar tendency for results between zero and nonzero between-class differences in slope could also be confirmed for the case of \(C=1\) (Tables 1, 5, and 6 in the supplemental online materials).
Shifts in correct marginal estimation rates at each level of agreement rates under different true numbers of classes, sample sizes, and degrees of separation. a True number of classes (true trajectory is linear). b Sample sizes (true trajectory is linear). c Degree of separation (true trajectory is linear). d True number of classes (true trajectory is quadratic). e Sample sizes (true trajectory is quadratic). f Degree of separation (true trajectory is quadratic)
In the main simulation, parameter estimates reported in Biesanz et al. (2004) were used to generate data. To confirm the generalizability of the results, we specified different factor means (i.e., \(\mu _{I}\), \(\mu _{S1}\) and \(\mu _{S2}\)) and (class-invariant) variance–covariance matrix \({\varvec{\Phi }}\), and performed a similar supplemental simulation. Details of the specified parameter values are provided on the first page of the supplemental online materials. Although results showed slightly higher correct estimation rates on average, similar tendencies (e.g., a dominant influence of r, followed by N and C on correct estimation rates, and smaller but nonzero impact of misspecification of the time-invariant error variances) were observed, indicating high generalizability of the present simulation results (see Tables 7 and 8 and Fig. 1 of the supplemental online materials for details).
We also compared the correct estimation rates between SEM Trees and LGCMMs, limited to conditions of \(N=100, 400, 1600\) and \(d=0.5, 1.5, 2.5\), using the hlme function from lcmm package to implement LGCMMs.Footnote 8 We generated simulation data by a similar procedure to the main simulation, and the Bayesian information criterion (BIC) was used to estimate the number of classes for LGCMMs.Footnote 9 For comparative purposes with SEM Trees, in \(C=1\) or \(C=2\) we compared values of BIC from LGCMMs that assume one or two classes to estimate the number of classes, while in \(C=4\) we compared LGCMMs that assume one, two, three or four classes. The correct marginal estimation rates for LGCMMs are shown in Table 9 of the supplemental online materials. From this table, LGCMMs showed much lower correct estimation rates, and large rates (e.g. more than 0.80) were observed only in the specific conditions of \(C=2\), \(N=1600\), and \(d=2.5\). Although levels of the manipulated factors are somewhat different from the simulation conducted in Usami et al. (2017), their research also showed similar levels of correct estimation rates in LGCMMs. The difference between SEM Trees and LGCMMs can be attributed to the relatively larger values of r specified for SEM Trees in the present simulation. Therefore, as expected from Table 1 and Fig. 1, if we can collect covariates whose agreement rates are larger than 0.7 (when class sizes are equal, this corresponds to expected correlations of .40 between true latent profiles—coded as 0 or 1 for two classes split from each node—and the observed covariates) in many cases SEM Trees can uncover population heterogeneity more precisely than can LGCMMs. However, due to the smaller power for uncovering population heterogeneity in LGCMMs, when \(C=1\), LGCMM could almost perfectly estimate the number of classes (Table 10 in the supplemental online materials).
3 Discussion
We performed a large-scale simulation study investigating the performance of SEM Trees in identifying classes, newly manipulating the number of time points and classes (nodes) using linear and quadratic LGCMs as template models. We also considered the influences of model misspecification regarding functional forms of latent growth curves and error variances. The results can be summarized as follows:
(1) In SEM Trees, correct estimation rates of the number of classes are most strongly related to the agreement rate of the covariate with its true latent profile, which is consistent with the former research of Usami et al. (2017).
(2) The number of true classes C, which is newly specified in the current simulation, has a serious negative impact on correct estimation of the number of classes. This point is exacerbated in the case of \(C>4\), which are likely in actual applications. Therefore, previous investigations might show overly optimistic results regarding the performance of SEM Trees in general.
(3) Influences of the total sample size and degree of separation (distances) among classes are also notable in correct estimation rates, whereas the number of time points had almost no relation to the average rates.
(4) SEM Trees was sensitive to the influence of model misspecification with (time-invariant) error variances, but its impact was relatively small and the misspecification of the true functional form of the latent trajectory was not influential. Thus, SEM Trees was not always sensitive to the influence of model misspecification, and its impact is different according to a complex function of the type of misspecification as well as the statistical properties of the template model (stability of parameters and frequency of estimation problems in general). Simulation results provide important insights about the utility of (bivariate) LGCM-based SEM Trees over (bivariate) LCS-based SEM Trees, namely that the former is a much more robust and stable approach under possible model misspecifications in growing trees.
(5) To achieve average correct estimation rates of 0.80 or more in the current simulation design, covariates that satisfy \(r=0.8\) or more are required when \(C=2\), whereas the required r increases to 0.95 when \(C=4\). Likewise, \(r=0.9\) is a minimal requirement if the sample size is as small as \(N=200\) or \(N=400\). However, the required r becomes as small as \(r=0.8\) and \(r=0.7\) if N becomes greater as \(N=800\) and \(N=1600\), respectively. When useful covariates are not gathered and r is as small as \(r=0.6\), achieving 0.80 or more average correct estimation rates seems to be very difficult, even in extreme cases such as simpler classification structures (i.e., \(C=2\)), large sample size (e.g., \(N=3200\)), and large degree of separation (i.e., \(d=2.5\)).
(6) From the supplemental simulation, the above conclusions are almost unchanged even when between-class differences in slope are present. Rather, in such cases correct estimation rates increase on average.
(7) If we can collect covariates whose agreement rates are larger than \(r=0.7\), in many cases SEM Trees can uncover population heterogeneity more precisely than LGCMMs.
The present simulation study has clarified the performance of SEM Trees, showing the dominant influences of agreement rates of the covariates as well as the sample size, and the true number of classes in detecting population heterogeneity in latent trajectories using an LGCM as the template model. SEM Trees detect classes according to covariates, so gathering useful covariates which can effectively explain the individual differences of growth parameters (e.g., intercepts and slopes) is essential in applying (LGCM-based) SEM Trees. To find such covariates, applying conditional LGCMs (where growth factors are regressed on covariates; e.g., Bollen and Curran 2006) must be a simple and useful strategy. If researchers expect that identifying or gathering such useful covariates will be difficult, it may be more efficient to apply unsupervised methods such as LGCMMs (which do not require covariates to extract classes). Comparing estimation results with those of SEM Trees must also be helpful to understand population heterogeneity from a different perspective. However, it should be noted that behavioral researchers have learned that estimation results, including the number of classes in LGCMMs, may be vulnerable to the influence of model misspecification. This indicates that robustness against model misspecification might be greatly different from SEM Trees, thus requiring additional future research that compare the performance of SEM Trees with LGCMMs under various kinds of model misspecification. In such future investigations, manipulation of the number of classes (with larger numbers than the current simulation), the stability of true latent trajectory (size of error variances), and methods for multiple testing (CV) should also be considered.
Given that in most cases, researchers may have uncertainty as to whether their covariates are informative, and may have more covariates than were tested in this simulation, there is a risk of detecting an erroneous classification structure due to an inflated type-I error. SEM Trees would manifest itself as splitting on covariates with no relation to the true latent profile, resulting in overfitting to the data. Especially with smaller sample sizes and larger numbers of covariates, researchers should be cautious in interpreting the resultant tree structure. As indicated by the present simulation, this point should be more critical when the true classification structure is complex (i.e., a large number of true classes). In that respect, although the computation burden is still heavy, using SEM Forests (Brandmaier et al. 2016) could be a useful alternative to address this problem, and more research is needed into the propensity for SEM Trees and SEM Forests to over-fit. From a technical aspect, additional future investigations should also include the development of more computationally efficient algorithms. On this point, The maximum likelihood-based method described in Merkle and Zeileis (2013) is potentially useful when it is extended for recursive partitioning, as this can greatly reduce computational burden in selecting split points (not requiring a new model to be estimated for each split point).
One potential extension of application of SEM Trees is to use a cross-lagged model as a template model for investigating population heterogeneity about reciprocal (or causal) effects between variables. Inferring reciprocal effects or causality between variables is a central aim in longitudinal research. However, while various longitudinal cross-lagged models have been proposed in various contexts with different backgrounds (e.g., Hamaker et al. 2015), similarities and differences of these models have been unclear, making it difficult for researchers to select a model that fits with the goal of their research. Using various cross-lagged models as template models and investigating the analysis results of SEM Trees under possible misspecification of the cross-lagged models chosen should provide an important finding that contributes to applied research.
We have to note that the importance of the issue of precisely estimating the number of classes might be largely different according to analytic purposes. Referring to the discussions in Bauer and Curran (2003) and Nagin and Tremblay (2005), the primary purposes of applying SEM Trees can be classified into two aspects: whether researchers aim to identify qualitatively distinct classes of individuals in the population of study, or if they just aim to approximate complex multivariate distributions with a small number of simpler component distributions. These two purposes of the model are theoretically quite distinct, but they are difficult to distinguish analytically (Bauer and Curran 2003). In the former case, estimating the true number of classes is a primary issue, and they may also need to identify “true” covariates that can explain qualitatively different classes. However, identifying such covariates is not necessarily important, because other covariates can identify true classification structures. In the latter case, estimating the true number of classes and identifying useful covariates might be trivial, because researchers typically aim to increase prediction accuracy. As a result, the extracted classes cannot usually be interpreted as true classification structures in the population. Although we primarily focused on the issue of estimating the number of classes in this simulation, future research should focus on situations where the primary purpose is prediction, not just correctly estimating the number of classes under various conditions that consider possible model misspecification. This should provide useful insights for researchers whose primary analytic purpose is prediction rather than estimating the number of classes.
The literature on longitudinal data design and analysis has been rapidly growing. SEM Trees and SEM Forests are powerful methods for relating informative covariates to a host of structural equation models, and allow researchers the ability to identify covariates that are important for understanding population heterogeneity. The results of our simulation study highlight conditions in which SEM Trees performs well, and conditions that result in lower rates of identifying the true group structure. Given that SEM Trees is a relatively new method, much more research is needed to evaluate the method. However, this should not detract from the use of the method. Instead, results should be interpreted with caution, both keeping the exploratory nature of the method in mind, as well as the uncertainty regarding how various conditions affect SEM Trees performance, especially when researchers are interested in estimating the true number of classes. We look forward to both more applied and simulation work moving into the future.
Notes
The LGCM can be expressed using a linear mixed model when time-invariant residual variances are assumed. This indicates that some of LGCM-based SEM Trees can be indirectly implemented using other tree methods based on linear mixed model. A recent development of Fokkema et al. (2017) proposed a generalized linear mixed-effects model tree algorithm to detect treatment-subgroup interactions. However, in this method the observed precision in detecting population heterogeneity has not been investigated under longitudinal design with the number of time points \(T\ge 3\), which is required in applying LGCM for identification.
Usami et al. (2017) conducted simulations in OpenMx because lavaan, which can provide faster computation, was not available in the semtree package at the start of simulations.
Because the number of time points (T) of child weight data is five, in the conditions of \(T=6\) and \(T=8\), error variances for \(t=6,7,8\) were specified as \(\psi _6^2=89.280\), \(\psi _7^2=91.947\), and \(\psi _8^2=89.336\) using predicted values based on estimated quadratic regression (i.e., predicted values \({\hat{\psi }}_t^2={\hat{\beta }}_0+{\hat{\beta }}_1t+{\hat{\beta }}_2t^2\) for \(t=6,7,8\) using the data of \(\psi _t^2\) reported at \(t=1,2,3,4,5\)).
When \(C=2\), the true latent profile of the first covariate \({\varvec{x_1}}\) is simply specified as \({\varvec{x_1}}=({\varvec{1^t_{N/2}}},{\varvec{0^t_{N/2}}})^t\). In contrast, when \(C=4\), where two covariates \({\varvec{x_1}}\) and \({\varvec{x_2}}\) are included in the dataset, true latent profiles in each class are specified as
$$\begin{aligned} \left( \begin{array}{c} {\varvec{x^t_1}} \\ {\varvec{x^t_2}} \end{array} \right) ^t =\left( \begin{array}{cccc} {\varvec{1^t_{N/4}}} &{} {\varvec{1^t_{N/4}}} &{} {\varvec{0^t_{N/4}}} &{}{\varvec{0^t_{N/4}}} \\ {\varvec{1^t_{N/4}}} &{} {\varvec{0^t_{N/4}}} &{} {\varvec{1^t_{N/4}}} &{}{\varvec{0^t_{N/4}}} \end{array} \right) ^t \end{aligned}$$Thus, in the population \({\varvec{x_1}}\) is the first covariate used to split the whole dataset into two nodes (one consisting of participants from the first and second classes, and the other from the third and fourth classes) whose distances are 2d, and \({\varvec{x_2}}\) is the second one to split these nodes, resulting in four child nodes (classes) whose distances between adjacent classes are d. In this simulation, we generated covariates so that both covariates have the same agreement rate r when \(C=4\) (i.e., \(r=r_1=r_2\)).
The Cross Validation (CV) method was not selected for multiple testing since it could not be applied to lavaan objects without errors. We confirmed that when selecting the “naive” method, simulation results were almost identical irrespective of the explicit specification of the Bonferroni method by “semtree.control()$bonferroni” in semtree.
It is obvious that underestimation of the number of classes never occurs when \(C=1\), while overestimation never occurs when \(C=2\) or \(C=4\) in the main simulation. Although there cannot be covariates that would split a parent node in the population when \(C=1\), we manipulated the agreement rate of dichotomous covariates r in this supplemental simulation. In this case, r can be interpreted as merely a proportion of dichotomous covariate that takes one.
We set two conditions of between-class differences in slope. Specifically, the slope mean for each class was specified using a random variable from a normal distribution \(N(6.988, 0.25^2\times 6.988^2)\) or \(N(6.988, 0.50^2\times 6.988^2)\) in each repetition. These conditions are equivalent to set the expected coefficients of variation (or relative standard deviation; the ratio of standard deviation of the specified slope mean (\(=0.25\times 6.988\) or \(0.50\times 6.988\)) to the original slope mean (\(=6.988\)) in each class) as 0.25 or 0.50 when randomly generating the slope means of classes in each repetition. We call these two conditions the intermediate and large between-class differences in slope in Tables 2 and 3 of the supplemental online materials, respectively.
Because hlme is originally a function for linear mixed models (rather than growth curve models), it implicitly assumes time-invariant error variances. This supplemental simulation thus assumes time-invariant error variances in the population, and the harmonic mean of (time-variant) residual variances (\(=20.294\)) is used for the parameter value. As a result, in this supplemental simulation we do not consider the presence of model misspecifications in error variances. In addition, the agreement rate of dichotomous covariates r was not manipulated, because LGCMMs do not require covariates to extract classes.
Regardless of which model selection procedure (e.g., the Akaike information criterion (AIC), sample-size-adjusted BIC) was used, Usami et al. (2017) found similar levels of correct estimation rates in their simulations. Thus, we did not use other model selection procedures here.
References
Bauer DJ, Curran PJ (2003) Distributional assumptions of growth mixture models: implications for overextraction of latent trajectory classes. Psychol Methods 8:338–363
Bauer DJ, Curran PJ (2004) The integration of continuous and discrete latent latent variable models: potential problems and promising opportunities. Psychol Methods 9:3–29
Berlin KS, Parra GR, Williams NA (2014) An introduction to latent variable mixture modeling (part 2): longitudinal latent class growth analysis and growth mixture models. J Pediatr Psychol 39:188–203
Biesanz JC, Deeb-Sossa N, Papadakis AA, Bollen KA, Curran PJ (2004) The role of coding time in estimating and interpreting growth curve models. Psychol Methods 9:30–52
Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick TR, Spies J, Estabrook R, Kenny S, Bates TC, Mehta P, Fox J (2011) OpenMx: multipurpose software for statistical modeling (version R package version 1.0.4), Virginia. http://openmx.psyc.virginia.edu
Bollen KA, Curran PJ (2006) Latent curve models: a structural equation approach. Wiley, Hoboken
Brandmaier AM, Oertzen Tv, McArdle JJ, Lindenberger U (2013) Structural equation model trees. Psychol Methods 18:71–86. https://doi.org/10.1037/a0030001
Brandmaier AM, Oertzen Tv, McArdle JJ, Lindenberger U (2014) Exploratory data mining with structural equation model trees. In: McArdle JJ, Ritschard G (eds) Contemporary issues in exploratory data mining in the behavioral sciences. Routledge, New York, pp 96–127
Brandmaier AM, Prindle JJ, McArdle JJ, Lindenberger U (2016) Theory-guided exploration with structural equation model forests. Psychol Methods 21:566–582
Brodley E, Utgoff PE (1995) Multivariate decision trees. Mach Learn 19:45–77
Fokkema M, Smits N, Zeileis A, Hothorn T, Kelderman H (2017) Detecting treatment-subgroup interactions in clustered data with generalized linear mixed-effects model trees. Behav Res Methods. https://doi.org/10.3758/s13428-017-0971-x (in press)
Genolini C, Falissard B (2010) KmL: K-means for longitudinal data. Comput Stat 25:317–332
Hamaker EL, Kuiper RM, Grasman RPPP (2015) A critique of the cross-lagged panel model. Psychol Method 20:102–116
Hayes T, Usami S, Jacobucci R, McArdle JJ (2015) Using classification and regression trees (CART) and random forests to analyze attrition: results from two simulations. Psychol Aging 30:911–929
Hayes T, McArdle JJ (2017) Should we impute or should we weight? examining the performance of two CART-based techniques for addressing missing data in small sample research with nonnormal variables. Comput Stat Data Anal 115:35–52
Henson JM, Reise SP, Kim KH (2007) Detecting mixtures from structural model differences using latent variable mixture modeling: A comparison of relative model fit statistics. Struct Equ Model 14:202–226
Jacobucci R, Grimm KJ, McArdle JJ (2017) A comparison of methods for uncovering sample heterogeneity: structural equation model trees and finite mixture models. Struct Equ Model 24:270–282
Leiby BE, Sammel MD, Ten Have TR, Lynch KG (2009) Identification of multivariate responders and non-responders by using Bayesian growth curve latent class models. J R Stat Soc Ser C 58:505–524
Little TD, Schnabel KU, Baumert J (eds) (2000) Modeling longitudinal and multiplegroup data. Practical issues, applied approaches, and specific examples. Lawrence Erlbaum Associates, Hillsdale
Martin DP (2015) Efficiently exploring mulilevel data with recursive partitioning. Unpublished doctoral dissertation, University of Virginia
Miller PJ, Lubke GH, McArtor DB, Bergeman CS (2015) Finding structure in data using multivariate tree boosting. arXiv:1511.02025
McArdle JJ (2009) Latent variable modeling of differences and changes with longitudinal data. Annu Rev Psychol 60:577–605
McArdle JJ, Nesselroade JR (2014) Longitudinal data analysis using structural equation models. American Psychological Association, Washington
McLachlan G, Peel DA (2000) Finite mixture models. Wiley, New York
Merkle EC, Zeileis A (2013) Tests of measurement invariance without subgroups: a generalization of classical methods. Psychometrika 78:59–82. https://doi.org/10.1007/s11336-012-9302-4
Meredith W, Tisak J (1984) On “Tuckerizing” curves. Presented at the annual meeting of the Psychometric Society, Santa Barbara, CA
Meredith W, Tisak J (1990) Latent curve analysis. Psychometrika 55:107–122
Morgan JN, Sonquist JA (1963) Ploblems in the analysis of survey data, and a proposal. J Am Stat Assoc 28:415–435
Nagin DS (1999) Analyzing developmental trajectories: a semi-parametric, group-based approach. Psychol Methods 4:139–157
Nagin DS, Land KC (1993) Age, criminal careers, and population heterogeneity: specification and estimation of a nonparametric, mixed Poisson model. Criminology 31:327–362
Nagin DS, Tremblay RE (1999) Trajectories of boys’ physical aggression, opposition, and hyperactivity on the path to physically violent and non violent juvenile delinquency. Child Dev 70:1181–1196
Nagin DS, Tremblay RE (2005) Developmental trajectory groups: fact or a useful statistical fiction? Criminology 43:873–904
Neelon B, Swamy Gk, Burgette LF, Miranda ML (2011) A Bayesian growth mixture model to examine maternal hypertension and birth outcomes. Stat Med 30:2721–2735
Preacher KJ, Zhang Z, Zyphur MJ (2016) Multilevel structural equation models for assessing moderation within and across levels of analysis. Psychol Methods 21:189–205
R Core Team (2016) R: a language and environment for statistical computing. R Foundation for statistical computing, Vienna, Austria. https://www.R-project.org/
Ram N, Grimm KJ (2009) Methods and measures: growth mixture modeling: a method for identifying differences in longitudinal change among unobserved groups. Int J Behav Dev 33:565–576
Rosseel Y (2012) lavaan: an R package for structural equation modeling. J Stat Softw 48(2):1–36
Sanchez G (2009) PATHMOX approach: segmentation trees in partial least squares path modeling. Unpublished doctoral dissertation, Universitat Politecnica de Catalunya, Catalonia, Spain
Sela RJ, Simonoff JS (2012) RE-EM trees: a data mining approach for longitudinal and clustered data. Mach Learn 86:169–207
Sonquist JA, Morgan JN (1964) The detection of interaction effects: a report on a computer program for the selection of optimal combinations of explanatory variables (No. 35). University of Michigan, Institute for Social Research, Survey Research Center, Ann Arbor
Todo N, Usami S (2016) Fitting unstructured finite mixture models in longitudinal design: a recommendation for model selection and estimation of the number of classes. Struct Equ Model 23:695–712
Usami S (2014a) Constrained \(k\)-means on cluster proportion and distances among clusters for longitudinal data analysis. Jpn Psychol Res 56:361–372
Usami S (2014b) Performances of information criteria for model selection in a latent growth curve mixture model. J Jpn Soc Comput Stat 27:17–48
Usami S, Hayes T, McArdle JJ (2015) On the mathematical relationship between latent change score model and autoregressive cross-lagged factor approaches: cautions for inferring causal relationship between variables. Multivar Behav Res 50:676–687
Usami S, Hayes T, McArdle JJ (2017) Fitting structural equation model trees and latent growth curve mixture models in longitudinal designs: the influence of model misspecification in estimating the number of classes. Struct Equ Model 24:585–598
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Usami, S., Jacobucci, R. & Hayes, T. The performance of latent growth curve model-based structural equation model trees to uncover population heterogeneity in growth trajectories. Comput Stat 34, 1–22 (2019). https://doi.org/10.1007/s00180-018-0815-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00180-018-0815-x