Abstract
Aims/Hypothesis
To determine if acute overexpression of peroxisome proliferator-activated receptor, gamma, coactivator 1 beta (Pgc-1β [also known as Ppargc1b]) in skeletal muscle improves insulin action in a rodent model of diet-induced insulin resistance.
Methods
Rats were fed either a low-fat or high-fat diet (HFD) for 4 weeks. In vivo electroporation was used to overexpress Pgc-1β in the tibialis cranialis (TC) and extensor digitorum longus (EDL) muscles. Downstream effects of Pgc-1β on markers of mitochondrial oxidative capacity, oxidative stress and muscle lipid levels were characterised. Insulin action was examined ex vivo using intact muscle strips and in vivo via a hyperinsulinaemic–euglycaemic clamp.
Results
Pgc-1β gene expression was increased >100% over basal levels. The levels of proteins involved in mitochondrial function, lipid metabolism and antioxidant defences, the activity of oxidative enzymes, and substrate oxidative capacity were all increased in muscles overexpressing Pgc-1β. In rats fed a HFD, increasing the levels of Pgc-1β partially ameliorated muscle insulin resistance, in association with decreased levels of long-chain acyl-CoAs (LCACoAs) and increased antioxidant defences.
Conclusions
Our data show that an increase in Pgc-1β expression in vivo activates a coordinated subset of genes that increase mitochondrial substrate oxidation, defend against oxidative stress and improve lipid-induced insulin resistance in skeletal muscle.
Similar content being viewed by others
Introduction
Insulin resistance is a key defect underlying obesity, type 2 diabetes and other metabolic diseases. The precise factors leading to development of insulin resistance are not completely resolved; however, there is substantial literature showing that accumulation of lipids within skeletal muscle is strongly associated with reductions in insulin sensitivity [1, 2]. In particular, lipid intermediates, such as diacylgycerols (DAG), ceramides and long-chain acyl-CoAs (LCACoAs) are thought to antagonise numerous intracellular pathways, ultimately leading to insulin resistance [1, 2]. One theory that has recently emerged implicates defective mitochondrial fatty acid oxidation as a causal factor in the build-up of lipid intermediates within skeletal muscle, and the development of insulin resistance. This theory is based on reports of defects in various markers of mitochondrial function in insulin-resistant skeletal muscle (reviewed by Savage et al. and by Kraegen and Cooney [1, 3]). However, despite the large number of studies suggesting a role for mitochondrial dysfunction in the development of insulin resistance, there are also an increasing number of reports showing a disconnection between these two variables. For example, we and others have shown that high-fat feeding enhances mitochondrial oxidative capacity in skeletal muscle, even though lipid accumulates and insulin resistance develops in this tissue [4, 5]. Furthermore, mice with genetically induced mitochondrial dysfunction in skeletal muscle are not insulin resistant, even when challenged with a high-fat diet (HFD) [6, 7]. By contrast, several studies have shown that enhancing substrate oxidation in muscle can reduce intramuscular lipid accumulation and improve insulin sensitivity [8, 9].
Another factor that has recently been implicated in the development of insulin resistance is oxidative stress [10–12]. Several groups, including our own, have demonstrated in cells and animals that insulin resistance is associated with excessive production of reactive oxygen species (ROS), particularly from the mitochondrial respiratory chain [10, 11]. Although the exact mechanism(s) by which elevated ROS levels cause insulin resistance are currently unknown, genetic and pharmacological interventions that attenuate excessive ROS production have been shown to partially ameliorate insulin resistance [10–12].
Given that excess lipid accumulation and oxidative stress are both associated with the pathogenesis of insulin resistance, strategies that enhance lipid oxidation and/or increase antioxidant defence capacity under high-fat-fed conditions should exert favourable effects on insulin sensitivity. In this regard the peroxisome proliferator-activated receptor, gamma, coactivator 1 (PGC1) family of transcriptional coactivators, which includes PGC-1α, PGC-1β and PGC1-related coactivator 1, are attractive targets. PGC-1α and PGC-1β have been the most extensively characterised proteins in this family, and a number of reports have shown that these two coactivators are both key regulators of mitochondrial oxidative capacity and the expression of several key antioxidant enzymes [13–16]. However, although PGC-1α and PGC-1β regulate a large number of overlapping genes, studies in rodent models and cell-based systems have also shown distinct effects of these proteins on a number of metabolic variables such as muscle fibre type [17, 18], sphingolipid profile [19] and functional properties of mitochondria [15].
Evidence of a potential role for defective PGC-1 signalling in metabolic disease has come from studies that show reductions in the expression of PGC-1α (also known as PPARGC1A) and PGC-1β (also known as PPARGC1B) in skeletal muscle of participants with insulin resistance and/or type 2 diabetes [20, 21]. However, studies that have manipulated the levels of PGC-1 proteins in mice have yielded unexpected and conflicting results. For example, skeletal muscle insulin action is not impaired in mice with whole body or muscle-specific deletion of PGC-1α or loss of function mutation of PGC-1β, despite these animals displaying the expected reduction in markers of mitochondrial metabolism [16, 22]. Furthermore, mice with muscle-specific overexpression of PGC-1α display a marked enhancement of oxidative characteristics in muscle, but are also insulin resistant owing to excessive fatty acid delivery to muscle or decreased Glut4 (also known as Slc2a4) expression [23, 24].
Whereas the above studies do not support a therapeutic role for activation of PGC-1 signalling, it should be noted that a number of complex adaptations may occur in mice owing to lifelong overexpression or ablation of a specific protein of interest. Indeed recent work from Bonen et al. has shown that acute overexpression of Pgc-1α in skeletal muscle of adult rats improves insulin action in both healthy and insulin-resistant muscles [25, 26]. In this study our aim was to determine the effect of acute overexpression of the related coactivator Pgc-1β on markers of mitochondrial function, oxidative stress and insulin action in muscle of high-fat-fed rats.
Methods
Animals
Male Wistar rats (∼250 g) were used for all experiments. Animals were obtained from the Animal Resources Centre (Perth, WA, Australia), and were maintained at 22 ± 0.5°C on a 12 h light–dark cycle. Rats were fed ad libitum for 4 weeks with either a standard rodent chow diet (∼10% energy from fat; Gordon’s Specialty Stock Feeds, Yanderra, NSW, Australia), or a HFD (45% of energy from fat [lard]) made in-house as described elsewhere [4]. All experimental procedures were approved by the Garvan Institute/St Vincent’s Hospital Animal Experimentation Ethics Committee and were in accordance with the National Health and Medical Research Council of Australia Guidelines on Animal Experimentation.
In vivo electrotransfer
Specific details of the electrotransfer procedure have been described previously [8, 27]. Animals were electroporated 2.5 weeks after commencement of their respective diets, and were killed 1.5 weeks after electroporation, unless otherwise stated. In brief, the murine Pgc-1β coding sequence [28] was cloned into a muscle-specific expression vector [27]. Control and test muscles were pretreated for 2 h with 90 units of hyaluronidase to break down components of the extracellular matrix, thus improving transfection efficiency [29]. The construct was resuspended in sterile saline (0.9% NaCl, 0.5 mg/ml) and tibialis cranialis (TC) and extensor digitorum longus (EDL) muscle received either six or three evenly spaced 50 μl injections, respectively. TC and EDL muscles from the contralateral limb received an equal volume of empty vector as a control. Both legs underwent an electroporation protocol consisting of one 800 V/cm, 100 ms pulse followed by four 80 V/cm, 100 ms pulses at 1 Hz.
Gene expression analysis
RNA was isolated from tissues using TRIzol reagent (Invitrogen, Mulgrave, VIC, Australia), according to the manufacturers’ instructions. After DNase digest, 0.65 μg of total RNA was reverse transcribed and analysed using a real-time qPCR 7900 system (Applied Biosystems, Foster City, CA, USA), using the standard curve method. Pgc-1β expression was detected using primers designed to cross-react with mouse and rat and relative gene expression was normalised to cyclophilin as an endogenous control (primer sequences are shown in the electronic supplementary material [ESM] Table 1).
Mitochondrial DNA copy number
Total DNA was isolated using a DNeasy kit (Qiagen, Mississauga, ON, USA), according to the manufacturers’ instructions. Relative mitochondrial DNA (mtDNA) copy number was determined by qPCR analysis of the ratio of mitochondrial-encoded and nuclear-encoded genes. Relative expression was determined using the qPCR ∆Ct standard curve method using 5 ng of template DNA. The primer pairs used were: (1) NADH dehydrogenase subunit 5 and solute carrier family 16 member 1 and (2) mitochondrial D-loop and β-actin (primer sequences are shown in ESM Table 1).
Western blot analysis
Muscle tissue (∼40 mg) was homogenised in ice-cold radioimmunoprecipitation assay (RIPA) buffer with protease inhibitors (50 mmol/l HEPES, 150 mmol/l NaCl, 5 mmol/l EDTA, 1% (wt/vol.) Nonidet NP-40, 0.5% (wt/vol.) sodium deoxycholate, 0.1% (wt/vol.) SDS, 10 mmol/l NaF, 1 mmol/l Na3VO4, 1 μg/ml leupeptin, 100 mg/ml phenylmethylsulfonyl fluoride and 2 mg/ml aprotinin). Lysates were resolved by SDS-PAGE and immunoblotted with antibodies against PGC-1α (Chemicon International, Temecula, CA, USA), mitochondrial respiratory chain subunits (complex I subunit NDUFB6, complex II FeS subunit, complex III Core2 subunit, complex V subunit alpha; MS601, Mitosciences, Eugene, OR, USA), carnitine palmitoyl transferase-1 (CPT-1; Alpha Diagnostic International, San Antonio, TX, USA), superoxide dismutase 1 and 2 (SOD-1 and SOD-2) and fatty acid translocase/CD36 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), uncoupling protein 3 (UCP3; Affinity Bioreagents, Golden, CO, USA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and porin (Cell Signaling Technology, Danvers, MA, USA), acyl-coA synthetase 1 (ACS1; a gift from P. Watkins at the Kennedy Krieger Institute, Baltimore, MD, USA), and GLUT4 (a gift from D. James, Garvan Institute of Medical Research, Sydney, WA, Australia). Immunolabelled bands were quantified by densitometry.
Substrate oxidation, mitochondrial respiration and oxidative enzyme activity in TC
To examine pyruvate and palmitate oxidation in control and transfected muscles, a 5% (wt/vol.) homogenate of fresh TC muscle was prepared in ice-cold homogenising buffer (250 mmol/l sucrose, 10 mmol/l Tris-HCl, 1 mmol/l EDTA, pH 7.4). Fifty microlitres of the homogenate was incubated in 450 μl prewarmed (30°C) oxidation medium (100 mmol/l sucrose, 10 mmol/l Tris-HCl, 5 mmol/l KH2PO4, 1 mmol/l MgCl2, 80 mmol/l KCl, 0.2 mmol/l EDTA, 1 mmol/l dithiothreitol, 2 mmol/l ATP, 0.3% fatty-acid free BSA, pH 7.4), supplemented with either 2 mmol/l carnitine, 0.05 mmol/l CoA, 0.1 mmol/l malate, 200 μmol/l palmitate, and 74 kBq/ml [1-14C]palmitic acid (GE Healthcare Life Sciences, Buckinghamshire, UK), or 2 mmol/l malate, 5 mmol/l pyruvate, and 14.8 kBq/ml [14C]pyruvic acid (GE Healthcare). Palmitate and pyruvate oxidation rates were determined as described elsewhere [4]. Mitochondria from control and test TC muscle were isolated as described in Turner et al. [30]. Respiration was measured polarographically using a Clark-type electrode (Strathkelvin Instruments, Glasgow, UK) at 30°C. The respiration medium contained 225 mmol/l mannitol, 75 mmol/l sucrose, 10 mmol/l Tris, 10 mmol/l K2HPO4, 0.1 mmol/l EDTA and 0.3% BSA, pH 7.4, and the substrate combinations used were 5 mmol/l pyruvate + 2 mmol/l malate, 10 mmol/l succinate + 4 μmol/l rotenone and 10 μmol/l palmitoyl-CoA+2 mmol/l l-carnitine + 2 mmol/l malate. State III respiration was initiated by adding 0.2 mmol/l ADP. The activity of citrate synthase (CS) and β-hydroxyacyl dehydrogenase (βHAD) were determined in TC muscle homogenates as previously described [4].
Ex vivo glucose uptake in EDL muscles
Control and Pgc-1β transfected muscles were dissected tendon-to-tendon into strips, and incubated in warmed (30°C), pregassed (95% O2:5% CO2, pH 7.4), modified Krebs–Henseleit buffer containing 5 mmol/l glucose, 10 mmol/l HEPES and 1% BSA. Following a 30 min preincubation period, muscle strips were transferred to a new vial containing the above medium plus 27.7 kBq/ml 2-deoxy[3H]glucose and 18.5 kBq/ml [14C]glucose (GE Healthcare) ± insulin (1 mU/ml) for 60 min. At the conclusion of the incubation period, muscle strips were rinsed in saline and dissolved in 1 mol/l KOH at 70°C. The extract was neutralised using 25% perchloric acid, and centrifuged at 13,000×g for 5 min at 4°C. The counts of phosphorylated deoxy[3H]glucose from this extract were used to calculate glucose uptake.
Hyperinsulinaemic–euglycaemic clamp
For hyperinsulinaemic–euglycaemic clamps in rats, double jugular cannulae were implanted 7 days before experiments. Animals (5 h fasted) were studied over 2 h in the conscious state (insulin infusion 0.25 units kg–1 h–1), with a bolus of 2-deoxy[3H]glucose tracer administered once plasma glucose levels reached steady state [8, 27]. At the conclusion of the clamp the TC and EDL muscles were rapidly dissected and freeze-clamped for determination of glucose uptake [8, 27].
Blood analyses
Plasma glucose was determined using an automated glucose analyser (YSI 2300; YSI, Yellow Springs, OH, USA). Insulin was assessed using a rat insulin ELISA (Linco, St Charles, MO, USA).
Lipid analysis
Lipids were extracted from muscle samples using the Folch method [31]. Total lipid extracts were separated into lipid classes using normal phase liquid chromatography and measured by evaporative light scattering detection [32]. The DAG and ceramide fractions were collected. For quantification of DAGs, the relevant fraction was infused into the QSTAR quadrupole time of flight mass spectrometer (Applied Biosystems/MDS Analytical Technologies, Toronto, ON, Canada), using the chip-based nanoelectrospray system Nanomate (Advion Biosciences, Ithaca, NY, USA). Positive electrospray precursor ion scanning mode was used [33]. Ceramide analysis was performed using reversed phase HPLC coupled to a triple quadrupole Quattro Premiere mass spectrometer (Waters, Milford, MA, USA). Total LCACoAs were measured with a fluorometric assay, as described by Antinozzi et al. [34].
Analysis of glutathione peroxidase (GPx) activity and lipid peroxidation
GPx activity was measured in TC muscle homogenates using the procedure described by Flohé and Günzler [35]. Oxidative modification of muscle lipids was determined spectrophotometrically as the amount of thiobarbituric acid-reactive substances (TBARS) formed [36]. Briefly, muscle was homogenised in RIPA buffer (described above) and proteins were precipitated with 10% trichloroacetic acid. Thiobarbituric acid (0.67% wt/vol.) was added to the supernatant fraction and samples were assayed at 532 nm after 10 min incubation at 100°C.
Statistics
All data are reported as means ± SE. Data were analysed with paired t test or two-way ANOVA. Post hoc analysis was conducted using a Bonferroni test together, where appropriate, with paired t test to specifically determine the effect of electroporation in each group. Statistical significance was accepted at p < 0.05.
Results
Timecourse of Pgc-1β overexpression in electroporated muscles
Pgc-1β gene expression in the TC muscle of chow-fed animals was increased approximately three-fold over basal at 4 days post-electroporation (Fig. 1a), and remained significantly elevated for up to 3 weeks. All subsequent experiments were conducted within the 8–16 day range, corresponding to an average increase in Pgc-1β mRNA of ∼2.5-fold (Fig. 1b). Interestingly, the HFD alone induced a significant (p < 0.05) increase in Pgc-1β expression, which was further elevated with electroporation (Fig. 1b).
Pgc-1β overexpression in electroporated TC muscles. (a) Time course of Pgc-1β expression in chow-fed animals at 4, 8, 12, 16 and 20 days after electroporation, n = 3–5 for each time point. (b) Average expression over basal at 8–16 days post-electroporation, n = 6 chow, n = 12 HFD. Data are means±SE. Specific effect of Pgc-1β (control vs test muscles); *p < 0.05, **p < 0.01, ‡ p = 0.07, § p < 0.05 main effect of diet, † p < 0.001 main effect of Pgc-1β. Grey and black bars represent control and Pgc-1β-overexpressing muscles, respectively
Effect of Pgc-1β overexpression on levels of metabolic proteins
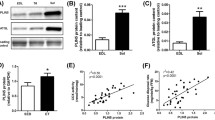
Significant upregulation (12–70%) of a number of proteins involved in mitochondrial function and lipid metabolism, including subunits of complex I and complex II of the mitochondrial respiratory chain, CPT-1, CD36, ACS and UCP3, was observed in TC muscles overexpressing Pgc-1β, compared with control muscles (Fig. 2a,b). There was also increased content of another well-known target of the PGC-1 proteins, GLUT4; however, the level of porin (an abundant mitochondrial outer membrane protein) was unchanged as a result of Pgc-1β overexpression (Fig. 2b). Despite significant increases in a number of mitochondrial proteins, we observed no difference in mtDNA content between control and Pgc-1β -overexpressing muscles, as assessed by the ratio of mtDNA to nuclear DNA using two different primer pairs (1.33 ± 0.16 vs 1.15 ± 0.26 AU for NADH dehydrogenase subunit 5: solute carrier family 16, member 1 and 0.97 ± 0.08 vs 0.83 ± 0.04 arbitrary units for mitochondria D-loop: β-actin for control vs Pgc-1β-expressing legs, respectively). Importantly, all of the above effects were specifically the result of increasing Pgc-1β expression, as there was no change in the content of PGC-1α (Fig. 2b).
Markers of mitochondrial metabolism in control and Pgc-1β-overexpressing TC muscles. (a) Representative immunoblots of proteins in chow-fed animal control (C) and test (T) muscles and (b) densitometric quantification of protein blots. For panels a and b, protein subunits examined in the mitochondrial complexes were as follows: complex I subunit NDUFB6, complex II FeS subunit, complex III Core2 subunit, complex V subunit alpha. Data are means±SE. *p < 0.05, **p < 0.01, ***p < 0.001 vs control. Dashed line represents corresponding control muscles (=1), and black bars represent Pgc-1β-overexpressing muscles. GAPDH, glyceraldehyde-3-phosphate dehydrogenase
Oxidative enzyme activity and substrate oxidation rates are increased in muscles overexpressing Pgc-1β
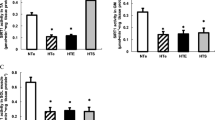
Pgc-1β overexpression increased (p < 0.01) the activity of CS and βHAD by ∼20% in chow-fed animals (Fig. 3a). Consistent with our previous findings [4] high-fat feeding increased the activity of CS (17%) and βHAD (28%) in control muscles, and Pgc-1β overexpression further enhanced this by 15% and 25%, respectively (Fig. 3a). To further elucidate the functional effects of Pgc-1β overexpression, we first examined pyruvate and palmitate oxidation rates in TC muscle homogenates. Pgc-1β overexpression increased the rate of pyruvate oxidation by ∼15% (p < 0.01, Fig. 3b), and palmitate oxidation by ∼25% (p < 0.001, Fig. 3c) in both chow and high-fat-fed animals. In mitochondria isolated from TC muscles of chow and fat-fed animals, Pgc-1β overexpression also increased ADP-stimulated respiration with pyruvate, succinate or palmitoyl-CoA as substrates, the latter again being above the adaptive response induced by an HFD (Fig. 3d–f).
Oxidative enzyme activity and substrate oxidation rates in control and Pgc-1β overexpressing TC muscles. (a) Enzyme activity of CS and βHAD, and oxidation rates of muscle homogenates incubated in medium containing (b) pyruvate or (c) palmitate. ADP-stimulated (state 3) respiration rates of mitochondria isolated from electroporated TC muscles incubated with (d) palmitoyl-CoA, (e) succinate and (f) pyruvate as substrates. Data are means±SE of n = 11–22 high-fat-fed, n = 6–12 chow-fed animals. Main effect of diet; ‡ p ≤ 0.05, ‡‡‡ p < 0.001, Main effect of Pgc-1β; † p ≤ 0.05, †† p < 0.01, ††† p < 0.001. Specific effect of electroporation (control vs Pgc-1β); *p < 0.05, **p < 0.01, ***p < 0.001. Grey and black bars represent control and Pgc-1β-overexpressing muscles, respectively
Pgc-1β overexpression in muscle ameliorates diet-induced insulin resistance
To determine if Pgc-1β overexpression in muscle of fat-fed rats improves diet-induced insulin resistance we examined insulin action under both ex vivo and in vivo conditions. High-fat feeding increased body mass and the mass of the epididymal and inguinal fat pads (Table 1). In EDL muscle strips from chow-fed animals, Pgc-1β overexpression did not alter the rate of insulin-stimulated glucose uptake (Fig. 4a). High-fat feeding induced marked insulin resistance in muscle (41% decrease in insulin-stimulated glucose uptake, p < 0.05 vs chow animals), whereas in Pgc-1β overexpressing EDL muscles from high-fat-fed animals, insulin action was almost completely restored to that observed in chow-fed animals (Fig. 4a).
Glucose uptake in control and Pgc-1β-overexpressing TC and EDL muscles. (a) Glucose uptake in EDL muscle strips under basal and insulin-stimulated conditions, data are means±SE of n = 9 high-fat-fed, n = 4 chow-fed animals. *p < 0.01 vs corresponding basal sample. Grey and black bars represent basal and insulin-stimulated conditions, respectively. Glucose uptake in (b) TC and (c) EDL muscles during a hyperinsulinaemic–euglycaemic clamp. The dotted line represents the average glucose uptake value in muscle from chow-fed animals. Data are means±SE for n = 10–15 animals. † p < 0.05 vs control HFD muscle. Grey and black bars represent control and Pgc-1β-overexpressing muscles, respectively
To assess the effect of Pgc-1β overexpression on insulin action in vivo, hyperinsulinaemic–euglycaemic clamps were performed in chow- and high-fat-fed animals. Whole body insulin resistance in the high-fat-fed animals was evident as a reduction in the glucose infusion rate during the clamp (Table 1). Insulin-stimulated glucose uptake was reduced in both the TC (–20%, p < 0.05) and EDL (–28%, p < 0.01) of high-fat-fed animals (Fig. 4b,c). In both muscle types, overexpression of Pgc-1β restored glucose uptake rates to levels comparable to the control leg of chow-fed animals (Fig. 4b,c).
Effect of Pgc-1β overexpression on muscle lipid levels
High-fat feeding increased the level of LCACoAs (+130%), ceramides (+25%) and DAG (+40%) in muscle (Fig. 5). Pgc-1β overexpression did not attenuate the HFD-induced increase in ceramide or DAG levels, but almost completely reduced LCACoA levels back to that observed in chow-fed animals (Fig. 5a).
Lipid content in control and Pgc-1β-overexpressing TC muscle. (a) LCACoA (b) ceramide and (c) DAG content in chow- and high-fat-fed animals. ***p < 0.001 main effect of diet, *p < 0.05 specific effect of Pgc-1β (control vs Pgc-1β). Grey and black bars represent control and Pgc-1β -overexpressing muscles, respectively
Effect of Pgc-1β overexpression on antioxidant defences
Pgc-1β overexpression increased the protein content of the antioxidant enzymes, SOD-1 and SOD-2 (Fig. 6a) and also increased (p < 0.01) the activity of GPx (1.72 ± 0.10 vs 2.15 ± 0.13 μmol min–1 g–1, control vs Pgc-1β, n = 11). As an index of oxidative damage we measured the level of lipid peroxidation (via TBARS) in muscle. High-fat feeding increased lipid peroxidation by ∼25% in TC muscles from control legs and, consistent with an enhanced capacity for ROS detoxification, Pgc-1β overexpression significantly reduced lipid peroxidation in both chow and fat-fed animals (Fig. 6c).
Markers of oxidative stress in Pgc-1β overexpressing TC muscle. (a) Representative immunoblots of antioxidant proteins SOD-1 and SOD-2 in control (C) and Pgc-1β test (T) muscles, and (b) densitometric quantification of proteins; **p < 0.01 control vs test leg, n = 9–12. (c) TBARS in TC muscles of chow- and high-fat-fed rats following hyperinsulinaemic–euglycaemic clamps. ‡ p < 0.01 main effect of diet, † p < 0.05 main effect of Pgc-1β. *p < 0.05 specific effect of electroporation (control vs Pgc-1β). Grey and black bars represent control and Pgc-1β overexpressing muscles, respectively
Discussion
The PGC-1 family of transcriptional coactivators are now recognised as important potential therapeutic targets for the treatment of many diseases [14, 37, 38]. In the present study we investigated whether acute, targeted overexpression of the transcriptional coactivator Pgc-1β could impact upon insulin action in rat skeletal muscle. We observed that overexpression of Pgc-1β was able to drive a transcriptional programme that increased the capacity for mitochondrial substrate oxidation, improved antioxidant defences and partially protected against HFD-induced insulin resistance.
Gain- and loss-of-function studies have shown that PGC-1β regulates the expression of genes involved in mitochondrial function and lipid metabolism [15, 16, 38, 39]. However, although much is known about the physiological stimuli that increase PGC-1α content in muscle, the in vivo regulation of PGC-1β is less well understood. Indeed, there is substantial disparity in the literature in this regard, with some studies reporting no alterations in PGC-1β levels with denervation, exercise or obesity [40, 41], whereas others have shown two- to threefold increases in PGC-1β expression in muscle with diverse stimuli, including insulin stimulation [21], exercise [42], bariatric surgery [43], dietary restriction [44] and treatment with the adipokine apelin [45]. In the present study we also observed a twofold increase in Pgc-1β in rat TC muscle following 4 weeks of an HFD. Acute overexpression of Pgc-1β by ∼2.5 fold resulted in increased content of mitochondrial respiratory chain subunits and proteins involved in lipid metabolism (e.g. CPT-1 and CD36). Furthermore, oxidative enzyme activity and the capacity to oxidise several different substrates were also enhanced in muscle overexpressing Pgc-1β. Interestingly, the Pgc-1β-induced enhancement in mitochondrial oxidative capacity occurred in the absence of any change in mtDNA and porin, suggesting that Pgc-1β interacts with a specific subset of transcription factors in vivo that, when activated, drive only part of the mitochondrial biogenic programme.
An enhanced capacity for mitochondrial substrate oxidation in skeletal muscle has been linked with improved insulin action [8, 9, 25, 26]. In the current study, rats fed an HFD displayed clear insulin resistance at the whole-body and muscle level. In both ex vivo assays in isolated EDL strips and in TC and EDL muscles during a hyperinsulinaemic–euglycaemic clamp, Pgc-1β was able to partially ameliorate this diet-induced insulin resistance. The observed improvements in insulin sensitivity are important considering that Pgc-1β was overexpressed only for 1–2 weeks of the 4-week high-fat feeding regime and with the electroporation technique, only ∼50% of muscle fibres within a given muscle are transfected [27]. Animals with whole body overexpression of PGC-1β display increased oxidative capacity in muscle and improved insulin sensitivity compared with wild-type controls [38]. However, two independent groups have shown that, despite improvements in mitochondrial capacity, mice with muscle-specific overexpression of PGC-1α exhibit insulin resistance, owing either to unexpected reductions in GLUT4 content or excessive lipid uptake into muscle [23, 24]. Whereas the findings from these transgenic animals have been disappointing, it appears that more modest increases in the level of the PGC-1 coactivators (e.g. two- to threefold in the current study and [25, 26]) avoid many of the unanticipated consequences of excessive transgene expression and result in beneficial effects with regards to insulin sensitivity.
Numerous studies have demonstrated that insulin resistance is linked with increased amounts of various deleterious lipid intermediates within skeletal muscle [1, 2]. Accumulation of LCACoA in muscle has been observed in association with insulin resistance in high-fat fed rats [46] and in rats and humans infused with lipid [47, 48]. LCACoA have been shown to inhibit hexokinase activity, thereby impairing glucose phosphorylation and reducing glucose uptake [49]. In the current study, high-fat feeding induced a greater than twofold increase in the levels of LCACoA within muscle and in Pgc-1β -transfected muscles from HFD animals, these levels were returned to levels close to that observed in chow controls. This reduction in LCACoA would potentially relieve the allosteric inhibition of hexokinase [49] and enhance insulin-stimulated glucose uptake; however, such changes in hexokinase activity cannot be directly verified by ex vivo measurements once the allosteric regulator (i.e. LCACoA) has been diluted by homogenisation. LCACoA levels could be potentially reduced if fatty acid uptake and/or activation were decreased; however, this is unlikely to be the reason for the current findings, as Pgc-1β overexpression increased the expression of the fatty acid transport protein CD36 and the enzyme responsible for fatty acid activation, ACS1. One plausible explanation for why Pgc-1β overexpression decreases LCACoA levels is that the observed enhancement of mitochondrial fatty acid capacity, above that induced by a HFD, leads to an increased channelling of lipid into oxidative pathways, thereby lowering the LCACoA pool.
Interestingly, although Pgc-1β overexpression resulted in a marked reduction in LCACoA levels in muscle from fat-fed animals, it did not prevent the diet-induced increases in ceramide and DAG levels. Recent work has suggested that in the context of insulin resistance, it is not only the total level of intracellular lipid intermediates that is important, but also their subcellular localisation. An increased proportion of reactive lipid species at the membrane is associated with insulin resistance and manipulations that reduce the level of these lipids improve insulin action [8, 24]. Whether Pgc-1β-induced alterations in the subcellular distribution of ceramides and DAG plays any role in the beneficial effects of this transcriptional coactivator on insulin action remains to be determined.
In addition to its ability to upregulate mitochondrial oxidative capacity, and reduce LCACoA levels, part of the beneficial effect of Pgc-1β on insulin action is probably related to an increase in antioxidant defences. A complex relationship exists between ROS production and insulin action. Intermittent, low-level ROS production has been reported to enhance insulin action via improvements in insulin signal transduction [50]. However, other recent studies have shown that excessive or persistent generation of ROS in mitochondria contributes to the pathogenesis of insulin resistance [10, 11]. The exact mechanism(s) by which excess ROS antagonises insulin action is currently unknown; however, this effect appears to be independent of changes in the canonical PI3K/Akt insulin-signalling pathway [10]. Importantly, it has been shown that pharmacological and genetic manipulations that alleviate oxidative stress lead to improvements in insulin sensitivity [10–12]. PGC-1α and PGC-1β have both been shown to regulate the production of a range of antioxidant proteins in a variety of cells [14–16]. Consistent with these reports, we observed a robust increase in the protein levels of SOD-1 and SOD-2 and in glutathione peroxidase activity in response to Pgc-1β overexpression in muscle. Importantly, this increase in antioxidant defence capacity resulting from Pgc-1β overexpression reduced the level of oxidative damage, as evidenced by decreased lipid peroxidation (TBARS) levels. Therefore, it is possible that increased detoxification of ROS is an additional mechanism that partially contributes to the protective effect of Pgc-1β against HFD-induced insulin resistance.
In summary, we have shown that overexpression of Pgc-1β is sufficient to improve diet-induced insulin resistance in skeletal muscle. This effect was associated with increased mitochondrial oxidative capacity, decreased levels of LCACoA and reduced oxidative stress. Our findings suggest agents that can activate Pgc-1β-dependent pathways in muscle may have therapeutic potential for the treatment of insulin resistance and type 2 diabetes.
Abbreviations
- ACS1:
-
Acyl-CoA synthetase1
- βHAD:
-
β-Hydroxyacyl dehydrogenase
- CPT-1:
-
Carnitine palmitoyl transferase-1
- CS:
-
Citrate synthase
- DAG:
-
Diacylglycerol
- EDL:
-
Extensor digitorum longus
- GPx:
-
Glutathione peroxidase
- HFD:
-
High-fat diet
- LCACoA:
-
Long-chain acyl-CoA
- mtDNA:
-
Mitochondrial DNA
- PGC:
-
Peroxisome proliferator-activated receptor, gamma, coactivator
- RIPA:
-
Radioimmunoprecipitation assay
- ROS:
-
Reactive oxygen species
- SOD:
-
Superoxide dismutase
- TBARS:
-
Thiobarbituric acid-reactive substances
- TC:
-
Tibialis cranialis
- UCP:
-
Uncoupling protein
References
Savage DB, Petersen KF, Shulman GI (2007) Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev 87:507–520
Kraegen EW, Cooney GJ (2008) Free fatty acids and skeletal muscle insulin resistance. Curr Opin Lipidol 19:235–241
Turner N, Heilbronn LK (2008) Is mitochondrial dysfunction a cause of insulin resistance? Trends Endocrinol Metab 19:324–330
Turner N, Bruce CR, Beale SM et al (2007) Excess lipid availability increases mitochondrial fatty acid oxidative capacity in muscle: evidence against a role for reduced fatty acid oxidation in lipid-induced insulin resistance in rodents. Diabetes 56:2085–2092
Hancock CR, Han DH, Chen M et al (2008) High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc Natl Acad Sci U S A 105(22):7815–7820
Pospisilik JA, Knauf C, Joza N et al (2007) Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell 131:476–491
Wredenberg A, Freyer C, Sandstrom ME et al (2006) Respiratory chain dysfunction in skeletal muscle does not cause insulin resistance. Biochem Biophys Res Commun 350:202–207
Bruce CR, Hoy AJ, Turner N et al (2009) Overexpression of carnitine palmitoyltransferase-1 in skeletal muscle is sufficient to enhance fatty acid oxidation and improve high fat diet-induced insulin resistance. Diabetes 58:550–558
Turner N, Hariharan K, TidAng J et al (2009) Enhancement of muscle mitochondrial oxidative capacity and alterations in insulin action are lipid species dependent: potent tissue-specific effects of medium-chain fatty acids. Diabetes 58:2547–2554
Hoehn KL, Salmon AB, Hohnen-Behrens C et al (2009) Insulin resistance is a cellular antioxidant defense mechanism. Proc Natl Acad Sci USA 106:17787–17792
Anderson EJ, Lustig ME, Boyle KE et al (2009) Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J Clin Invest. doi:10.1172/JCI37048
Houstis N, Rosen ED, Lander ES (2006) Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature 440:944–948
Arany Z (2008) PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr Opin Genet Dev 18:426–434
St-Pierre J, Drori S, Uldry M et al (2006) Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127:397–408
St-Pierre J, Lin J, Krauss S et al (2003) Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem 278:26597–26603
Vianna CR, Huntgeburth M, Coppari R et al (2006) Hypomorphic mutation of PGC-1beta causes mitochondrial dysfunction and liver insulin resistance. Cell Metab 4:453–464
Lin J, Wu H, Tarr PT et al (2002) Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418:797–801
Arany Z, Lebrasseur N, Morris C et al (2007) The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab 5:35–46
Espinoza DO, Boros LG, Crunkhorn S, Gami H, Patti ME (2010) Dual modulation of both lipid oxidation and synthesis by peroxisome proliferator-activated receptor-gamma coactivator-1alpha and -1beta in cultured myotubes. FASEB J 24:1003–1014
Patti ME, Butte AJ, Crunkhorn S et al (2003) Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci USA 100:8466–8471
Ling C, Poulsen P, Carlsson E et al (2004) Multiple environmental and genetic factors influence skeletal muscle PGC-1alpha and PGC-1beta gene expression in twins. J Clin Invest 114:1518–1526
Handschin C, Choi CS, Chin S et al (2007) Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest 117:3463–3474
Miura S, Kai Y, Ono M, Ezaki O (2003) Overexpression of peroxisome proliferator-activated receptor gamma coactivator-1alpha down-regulates GLUT4 mRNA in skeletal muscles. J Biol Chem 278:31385–31390
Choi CS, Befroy DE, Codella R et al (2008) Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc Natl Acad Sci USA 105:19926–19931
Benton CR, Nickerson JG, Lally J et al (2008) Modest PGC-1alpha overexpression in muscle in vivo is sufficient to increase insulin sensitivity and palmitate oxidation in subsarcolemmal, not intermyofibrillar, mitochondria. J Biol Chem 283:4228–4240
Benton CR, Holloway GP, Han XX et al (2010) Increased levels of peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PGC-1alpha) improve lipid utilisation, insulin signalling and glucose transport in skeletal muscle of lean and insulin-resistant obese Zucker rats. Diabetologia 53:2008–2019
Cleasby ME, Davey JR, Reinten TA et al (2005) Acute bidirectional manipulation of muscle glucose uptake by in vivo electrotransfer of constructs targeting glucose transporter genes. Diabetes 54:2702–2711
Lelliott CJ, Ljungberg A, Ahnmark A et al (2007) Hepatic PGC-1beta overexpression induces combined hyperlipidemia and modulates the response to PPARalpha activation. Arterioscler Thromb Vasc Biol 27:2707–2713
McMahon JM, Signori E, Wells KE, Fazio VM, Wells DJ (2001) Optimisation of electrotransfer of plasmid into skeletal muscle by pretreatment with hyaluronidase—increased expression with reduced muscle damage. Gene Ther 8:1264–1270
Turner N, Li JY, Gosby A et al (2008) Berberine and its more biologically available derivative, dihydroberberine, inhibit mitochondrial respiratory complex I: a mechanism for the action of berberine to activate AMP-activated protein kinase and improve insulin action. Diabetes 57:1414–1418
Folch J, Lees M, Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226:497–509
Homan R, Anderson MK (1998) Rapid separation and quantitation of combined neutral and polar lipid classes by high-performance liquid chromatography and evaporative light-scattering mass detection. J Chromatogr B Biomed Sci Appl 708:21–26
Ejsing CS, Sampaio JL, Surendranath V et al (2009) Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA 106:2136–2141
Antinozzi PA, Segall L, Prentki M, McGarry JD, Newgard CB (1998) Molecular or pharmacologic perturbation of the link between glucose and lipid metabolism is without effect on glucose-stimulated insulin secretion. A re-evaluation of the long-chain acyl-CoA hypothesis. J Biol Chem 273:16146–16154
Flohe L, Gunzler WA (1984) Assays of glutathione peroxidase. Methods Enzymol 105:114–121
Ohkawa H, Ohishi N, Yagi K (1979) Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal Biochem 95:351–358
Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT (2009) Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci USA 106:20405–20410
Kamei Y, Ohizumi H, Fujitani Y et al (2003) PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci USA 100:12378–12383
Lelliott CJ, Medina-Gomez G, Petrovic N et al (2006) Ablation of PGC-1beta results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol 4:e369
Koves TR, Li P, An J et al (2005) Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem 280:33588–33598
Holloway GP, Perry CG, Thrush AB et al (2008) PGC-1alpha’s relationship with skeletal muscle palmitate oxidation is not present with obesity despite maintained PGC-1alpha and PGC-1beta protein. Am J Physiol Endocrinol Metab 294:E1060–1069
Ristow M, Zarse K, Oberbach A et al (2009) Antioxidants prevent health-promoting effects of physical exercise in humans. Proc Natl Acad Sci USA 106:8665–8670
Hernandez-Alvarez MI, Chiellini C, Manco M et al (2009) Genes involved in mitochondrial biogenesis/function are induced in response to bilio-pancreatic diversion in morbidly obese individuals with normal glucose tolerance but not in type 2 diabetic patients. Diabetologia 52:1618–1627
Oita RC, Mazzatti DJ, Lim FL, Powell JR, Merry BJ (2009) Whole-genome microarray analysis identifies up-regulation of Nr4a nuclear receptors in muscle and liver from diet-restricted rats. Mech Ageing Dev 130:240–247
Frier BC, Williams DB, Wright DC (2009) The effects of apelin treatment on skeletal muscle mitochondrial content. Am J Physiol Regul Integr Comp Physiol 297:R1761–1768
Ellis BA, Poynten A, Lowy AJ et al (2000) Long-chain acyl-CoA esters as indicators of lipid metabolism and insulin sensitivity in rat and human muscle. Am J Physiol Endocrinol Metab 279:E554–560
Hoy AJ, Brandon AE, Turner N et al (2009) Lipid and insulin infusion-induced skeletal muscle insulin resistance is likely due to metabolic feedback and not changes in IRS-1, Akt, or AS160 phosphorylation. Am J Physiol Endocrinol Metab 297:E67–75
Tsintzas K, Chokkalingam K, Jewell K, Norton L, Macdonald IA, Constantin-Teodosiu D (2007) Elevated free fatty acids attenuate the insulin-induced suppression of PDK4 gene expression in human skeletal muscle: potential role of intramuscular long-chain acyl-coenzyme A. J Clin Endocrinol Metab 92:3967–3972
Thompson AL, Cooney GJ (2000) Acyl-CoA inhibition of hexokinase in rat and human skeletal muscle is a potential mechanism of lipid-induced insulin resistance. Diabetes 49:1761–1765
Loh K, Deng H, Fukushima A et al (2009) Reactive oxygen species enhance insulin sensitivity. Cell Metab 10:260–272
Acknowledgements
L.E. Wright and J. Reznick are supported by University of New South Wales and Australian Postgraduate Awards, respectively. A.J. Hoy is supported by a Biomedical Australian Training Fellowship, N. Turner by a Career Development Award and G. J. Cooney by a Research Fellowship from the National Health and Medical Research Council of Australia. The excellent technical assistance of S. Beale and S. Leslie of the Garvan Institute of Medical Research is gratefully acknowledged. We thank J. Cantley and D. Saunders at the Garvan Institute of Medical Research for critically reviewing this manuscript. We also thank the Biological Testing Facility at the Garvan Institute for assistance with animal care.
Duality of Interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Table 1
Primer sequences for qPCR (PDF 20 kb)
Rights and permissions
About this article
Cite this article
Wright, L.E., Brandon, A.E., Hoy, A.J. et al. Amelioration of lipid-induced insulin resistance in rat skeletal muscle by overexpression of Pgc-1β involves reductions in long-chain acyl-CoA levels and oxidative stress. Diabetologia 54, 1417–1426 (2011). https://doi.org/10.1007/s00125-011-2068-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-011-2068-x