Abstract
Aims/hypothesis
The rs1990760 polymorphism (Ala946Thr) of interferon induced with helicase C domain 1 (IFIH1) has been proposed to associate with type 1 diabetes. In this study, association between IFIH1 Ala946Thr and type 1 diabetes was investigated in two distinct white populations, the Hungarians and Finns.
Methods
The rs1990760 polymorphism was genotyped in 757/509 Hungarian/Finnish childhood-onset cases, 499/250 Hungarian/Finnish control individuals and in 529/924 Hungarian/Finnish nuclear family trios. Disease association was tested using case–control and family-based approaches. A meta-analysis of data from 9,546 cases and 11,000 controls was also performed.
Results
In the Hungarian dataset, the A allele was significantly more frequent among cases than among controls (OR 1.29, 95% CI 1.10–1.52; p = 0.002). Combined analysis of Hungarian and Finnish datasets revealed a strong disease association (OR 1.235, 95% CI 1.083–1.408; p = 0.002). Furthermore, the A allele was significantly overtransmitted in both family trio datasets (p = 0.017 in Hungarians; p = 0.007 in Finns). The A allele was increased in Hungarian vs Finnish cases (64.9% vs 60.8% in Finns; p = 0.003). The meta-analysis yielded a significant effect for IFIH1 rs1990760 A allele on type 1 diabetes risk (OR 1.176, 95% CI 1.130–1.225; p = 5.3 × 10−15) with significant heterogeneity between effect sizes across the studied populations (p = 0.023).
Conclusions/interpretation
This study represents the first independent confirmation of the association between type 1 diabetes and the IFIH1 gene in Hungarian and Finnish populations. Summarising the data published so far, a clear association between the Ala946Thr polymorphism and type 1 diabetes was detected, with an apparent difference in the contribution to disease susceptibility in different populations of European ancestry.
Similar content being viewed by others
Introduction
Type 1 diabetes is a chronic autoimmune disease caused by the interplay of triggering environmental factors and genetic susceptibility [1].
Recently, a non-synonymous single nucleotide polymorphism (rs1990760, resulting in an adenine (A)/guanine (G) nucleotide and alanine [Ala] 946 threonine [Thr] amino acid change) in exon 15 of the gene encoding interferon induced with helicase C domain 1 (IFIH1, 2q24.3) showed an association with type 1 diabetes [2]. IFIH1 is a cytosolic viral recognition receptor, and may provide a link between viral infections and type 1 diabetes.
Since allelic frequencies of disease variants differ across populations, their contributions to disease manifestation may vary. It is likely that certain variants with permissive effects on environmental disease triggers contribute in different degrees to disease aetiology in various ethno-geographical settings.
Thus, in our study we aimed to assess the association between the IFIH1 Ala946Thr polymorphism and type 1 diabetes in two distinct populations of European origin, the Hungarians and the Finns. Hungarians are considered a medium-incidence population for type 1 diabetes with a mixed genetic background [3], whereas Finland has the highest disease incidence in the world and its population is relatively homogeneous [4]. Investigating these two well-characterised populations is of special interest as, besides their different genetic architecture, environmental factors and socioeconomic conditions vary greatly between the two countries. Recently, we have shown that enterovirus infections in early childhood are more common in Hungary than in Finland [5]. Therefore, our study was tailored to provide a deeper insight into the genetics of type 1 diabetes in populations with different frequencies of enteroviral infection. In addition, because of the relatively modest effect conferred by the IFIH1 Ala946Thr variant, we conducted a meta-analysis of the association studies published so far [2, 6, 7], including our new datasets, to address the contribution of IFIH1 rs1990760 polymorphism to type 1 diabetes. Furthermore, we examined the heterogeneity between the populations studied.
Methods
Participants
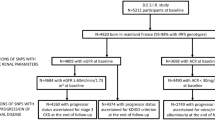
In the case–control study, Hungarian (n = 757) and Finnish (n = 509) patients with type 1 diabetes diagnosed under 15 years of age were included. The Hungarian cases were recruited from diabetes outpatient clinics all over Hungary, through the HUNT1DGENES programme (www.diabet.hu/diabetesgenetics, accessed 10 June 2009). The Finnish cases were collected from Turku and Helsinki regions through the Finnish Childhood Diabetes Registry. Healthy Hungarian blood donors (n = 499) were recruited as controls. The Finnish control group comprised geographically matched consecutive healthy newborns (n = 250).
For the family-based association study, 529 Hungarian unrelated parent–child trios (1587 individuals in total) were obtained from the HUNT1DGENES study. From the Finnish Childhood Diabetes Registry 924 family trios were collected.
Informed consent was obtained from participants. The study was approved by the national Ethics Committees and was conducted according to the principles of the Declaration of Helsinki.
Genotyping
The rs1990760 polymorphism was typed using the Taqman assay (Applied Biosystems, Foster City, CA, USA) or by iPlex Gold (Sequenom, San Diego, CA, USA). Genotyping accuracy was 100% as identified by fluorescence-based capillary DNA sequencing of 194 samples, representing all three genotypes in similar proportions (MegaBace1000, GE Healthcare, Chalfont St Giles, UK). No deviation from the Hardy–Weinberg equilibrium was found in any group.
Statistical analysis
For the case–control data the χ 2 test or Cochran–Mantel–Haenszel statistics were used and ORs were calculated. The statistical power to detect a genetic association at the originally reported OR of 1.18 [2] (α = 0.05) was 68.7% for the combined Hungarian and Finnish case–control series. The family data were analysed using the transmission-disequilibrium test (TDT). Meta-analysis assuming fixed effects was performed using MIX software [8], under the allelic genetic model. Between-study heterogeneity was assessed by Cochran’s Q test.
Results
In the Hungarian case–control dataset, the AA genotype of IFIH1 rs1990760 polymorphism was more frequent in cases than in controls (p = 0.011; Table 1). Accordingly, the A allele was significantly more common in cases than in controls (OR 1.291, 95% CI 1.096–1.521; p = 0.002). In the Finnish case–control dataset, we observed a similar difference in the genotype distribution between cases and controls; however, it did not reach significance (Table 1). Importantly, combined analysis of the Hungarian and Finnish case–control datasets showed a significant disease association of the IFIH1 rs1990760 polymorphism (for the A allele: OR 1.235, 95% CI 1.083–1.408; p = 0.002; Table 1).
In the Hungarian family trio data, 12 Mendelian errors were detected and these families were excluded from the final analysis. Of the remaining 517 families, 480 families had at least one heterozygous parent, and were therefore informative for TDT. The minor allele frequency of the G allele was 0.366, while the frequency of the A allele was 0.634. The A allele was significantly overtransmitted to affected offspring (266 transmissions and 214 non-transmissions, p = 0.017).
The Finnish family trio data contained no Mendelian errors, and of the 924 nuclear family trios 899 were informative for TDT. Minor G allele frequency was 0.398, whereas the major A allele frequency was 0.602. In a pattern similar to that in the Hungarians, the A allele was overtransmitted in the high-diabetes-incidence Finns with 490 transmissions and 409 non-transmissions (p = 0.007).
The comparison of allelic frequencies among all type 1 diabetes affected individuals revealed that the A allele frequency was higher among Hungarians than among Finns (64.9% vs 60.8%, p = 0.003), while no difference was found in controls (58.2% in both groups). We also observed that the genotype frequency of the disease-associated A allele carriers (AA+AG) between cases and controls in the two populations showed a significant difference (p = 0.003). The AG genotype was observed to be more frequent in Finnish cases (46.6% vs 43.5% in Hungarians), whereas among Hungarians the AG genotype was more frequent among controls (47.1% vs 42.0% in Finns; p = 0.001). However, there was no marked heterogeneity between the two populations (Cochran’s Q test, p = 0.379).
The meta-analysis of studies published so far, which comprised a total of 9,546 cases and 11,000 controls of European ancestry, yielded a modest but significant effect for the IFIH1 rs1990760 A allele on type 1 diabetes risk (OR 1.176, 95% CI 1.130–1.225; p = 5.3 × 10−15; Fig. 1). There was significant heterogeneity between effect sizes across the studied populations (Cochran’s Q test, p = 0.023).
Cumulative effect of IFIH1 rs1990760 polymorphism on the risk of type 1 diabetes in 9,546 cases and 11,000 controls using an allelic model of inheritance. Six populations of European ancestry from distinct geographical locations were included in the meta-analysis. A significant heterogeneity was observed across studies (Cochran’s Q test p = 0.023). Studies are ordered by publication date. ORs and 95% confidence intervals are shown. Sizes of OR symbols are proportional to the study sample size. T1D, type 1 diabetes
Discussion
The IFIH1 gene was originally found to confer susceptibility to type 1 diabetes in a whole-genome association scan [2]. Later, a French–Canadian family study indicated a trend of association; however, no significant effect was confirmed [9]. Furthermore, a US study reported an association between the IFIH1 rs1990760 and type 1 diabetes in the population of Georgia (state), USA; however, the authors could not replicate this finding in a population from the state of Colorado, USA [6]. In addition, a very recent Belgian study reported no association [7], whereas another report by Nejentsev et al. suggested that multiple additional rare variants of IFIH1 may contribute to disease susceptibility [10]. Therefore, the confirmation of the disease association of IFIH1 rs1990760 was awaited, especially in Europeans.
Here we showed that the IFIH1 Ala946Thr polymorphism is associated with type 1 diabetes in two large population samples of European origin: the very-high-incidence Finnish population and the medium-incidence Hungarian ethnic group. The association was clear both in the Hungarian case–control dataset and in the combined Finnish–Hungarian dataset. The association in the Finnish case–control series was not significant, but this could be partly due to reduced power. The family-based association study further confirmed the disease association in both populations.
Interestingly, we found that although the allele frequencies in the two background populations were identical, the frequency of the susceptibility A allele was higher among Hungarian cases compared with Finnish patients. These data suggest that the IFIH1 rs1990760 polymorphism may have a stronger effect in the Hungarian population, where early childhood infections are more common [5]. This is interesting as IFIH1 is a cytosolic viral sensor that, upon interaction with intracellular double-stranded RNA, induces production of antiviral cytokines and may induce apoptosis. Thus, IFIH1 may provide a link between the enterovirus infections and type 1 diabetes. However, comparing our Hungarian and Finnish cohorts no statistical evidence was found for different effect sizes of the IFIH1 polymorphism. Of note, our study was underpowered to show heterogeneity.
Nevertheless, to address the contribution of IFIH1 rs1990760 polymorphism to type 1 diabetes and to assess the heterogeneity between populations of European ancestry, we conducted a meta-analysis on previously published studies [2, 6, 7] including ours, using allelic genetic model. In this meta-analysis convincing statistical evidence was provided for an influence of the IFIH1 rs1990760 A allele on type 1 diabetes risk. Interestingly, a significant heterogeneity was found between effect sizes across the studied populations. This may indicate that the effect of IFIH1 rs1990760 polymorphism varies in different populations with distinct environmental influence.
In summary, this study represents the first independent confirmation of the association between type 1 diabetes and the IFIH1 gene in two well-defined European populations: the medium-incidence Hungarians and the Finns, who have the highest disease incidence in the world. A meta-analysis conducted in six different populations of European ancestry showed a clear association between the IFIH1 Ala946Thr polymorphism and type 1 diabetes, with an apparent difference in the contribution to disease susceptibility in distinct geographical locations, emphasising the role of environmental factors in the pathogenesis of type 1 diabetes.
Abbreviations
- A:
-
Adenine
- Ala:
-
Alanine
- G:
-
Guanine
- IFIH1:
-
Interferon induced with helicase C domain 1
- TDT:
-
Transmission-disequilibrium test
- Thr:
-
Threonine
References
Knip M, Veijola R, Virtanen SM, Hyoty H, Vaarala O, Akerblom HK (2005) Environmental triggers and determinants of type 1 diabetes. Diabetes 54(Suppl 2):S125–S136
Smyth DJ, Cooper JD, Bailey R et al (2006) A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 38:617–619
Hermann R, Bartsocas CS, Soltesz G et al (2004) Genetic screening for individuals at high risk for type 1 diabetes in the general population using HLA Class II alleles as disease markers. A comparison between three European populations with variable rates of disease incidence. Diabetes Metab Res Rev 20:322–329
DIAMOND Project Group (2006) Incidence and trends of childhood type 1 diabetes worldwide 1990–1999. Diabet Med 23:857–866
Viskari H, Ludvigsson J, Uibo R et al (2004) Relationship between the incidence of type 1 diabetes and enterovirus infections in different European populations: results from the EPIVIR project. J Med Virol 72:610–617
Liu S, Wang H, Jin Y et al (2009) IFIH1 polymorphisms are significantly associated with type 1 diabetes and IFIH1 gene expression in peripheral blood mononuclear cells. Hum Mol Genet 18:358–365
Aminkeng F, Van Autreve JE, Weets I et al (2009) IFIH1 gene polymorphisms in type 1 diabetes: genetic association analysis and genotype–phenotype correlation in the Belgian population. Hum Immunol 70:706–710
Bax L, Yu LM, Ikeda N, Tsuruta H, Moons KGM (2006) Development and validation of MIX: comprehensive free software for meta-analysis of causal research data. BMC Med Res Methodol 6:50
Qu HQ, Marchand L, Grabs R, Polychronakos C (2008) The association between the IFIH1 locus and type 1 diabetes. Diabetologia 51:473–475
Nejentsev S, Walker N, Riches D, Egholm M, Todd JA (2009) Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324:387–389
Acknowledgements
The authors are grateful to A. Doria for helpful discussions on statistical analysis, and to the clinical investigators of the HUNT1DGENES programme for their contribution. This study was supported by the Oszkár Asbóth Program of Hungarian NORT; TÁMOP-4.2.2-08/01/KMR-2008-2004; the Hungarian Diabetes Association; ETT 554/2006; the Academy of Finland, Varsinais Suomen Kultuurirahasto and the Sigrid Juselius Foundation. The technical assistance of B. Kovács, Z. Szabó and P. Nurmi and the support from the Hungarian National Blood Service are gratefully acknowledged.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
A. Jermendy and I. Szatmári contributed equally to this study.
The members of the Finnish Paediatric Diabetes Registry are listed in Laine AP, Holmberg H, Nilsson A et al (2007) Two insulin gene single nucleotide polymorphisms associated with type 1 diabetes risk in the Finnish and Swedish populations. Dis Markers 23:139–145.
Appendix
Appendix
Clinical investigators of the HUNT1DGENES programme: M. Drexler, Z. Almássy, T. Niederland, M. Balogh, G. Soltész, É. Erhardt, L. Barkai, E. Felszeghy, L. Blatniczky, Z. Nagyszakáll, É. Hosszú, I. Szabó, K. Kocsis, É. Gajzer, M. Dósa, B. Godó, P. Szűts, H. K. Nagy, T. Tóth, G. Fekete, M. Buzogány, Z. Karádi, I. Rippl, I. Kántor and I. Petheő.
Rights and permissions
About this article
Cite this article
Jermendy, A., Szatmári, I., Laine, A.P. et al. The interferon-induced helicase IFIH1 Ala946Thr polymorphism is associated with type 1 diabetes in both the high-incidence Finnish and the medium-incidence Hungarian populations. Diabetologia 53, 98–102 (2010). https://doi.org/10.1007/s00125-009-1561-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-009-1561-y