Abstract
Aim/hypothesis
Recent studies have demonstrated relationships between circadian clock function and the development of metabolic diseases such as type 2 diabetes. We investigated whether the peripheral circadian clock is impaired in patients with type 2 diabetes.
Methods
Peripheral leucocytes were obtained from eight patients with diabetes and six comparatively young non-diabetic volunteers at 09:00, 15:00, 21:00 and 03:00 hours (study 1) and from 12 male patients with diabetes and 14 age-matched men at 09:00 hours (study 2). Transcript levels of clock genes (CLOCK, BMAL1 [also known as ARNTL], PER1, PER2, PER3 and CRY1) were determined by real-time quantitative PCR.
Results
In study 1, mRNA expression patterns of BMAL1, PER1, PER2 and PER3 exhibited 24 h rhythmicity in the leucocytes of all 14 individuals. The expression levels of these mRNAs were significantly (p < 0.05) lower in patients with diabetes than in non-diabetic individuals at one or more time points. Moreover, the amplitudes of mRNA expression rhythms of PER1 and PER3 genes tended to diminish in patients with diabetes. In study 2, leucocytes obtained from patients with diabetes expressed significantly (p < 0.05) lower transcript levels of BMAL1, PER1 and PER3 compared with leucocytes from control individuals, and transcript expression was inversely correlated with HbA1c levels (ρ = −0.47 to −0.55, p < 0.05).
Conclusions/interpretation
These results suggest that rhythmic mRNA expression of clock genes is dampened in peripheral leucocytes of patients with type 2 diabetes. The impairment of the circadian clock appears to be closely associated with the pathophysiology of type 2 diabetes in humans.
Similar content being viewed by others
Introduction
The circadian system is responsible for regulating a variety of physiological and behavioural processes, including feeding behaviour and energy metabolism [1, 2]. Recent studies revealed that the circadian clock system consists essentially of a set of clock genes [1, 2]. The circadian clock resides in the hypothalamic suprachiasmatic nucleus (SCN), which is recognised as being the master clock, and the same clock exists also in almost all peripheral tissues, including liver, heart, kidney [3–5] and leucocytes [6–8]. Although the SCN is not essential for driving peripheral oscillations, it appears to coordinate peripheral clocks [5].
In mammals, rhythmic transcriptional enhancement by two basic helix–loop–helix transcription factors, clock homologue (mouse) (CLOCK) and brain and muscle Arnt-like protein 1 (BMAL1), provides the basic drive for the intracellular circadian clock (Electronic supplementary material [ESM] Fig. 1) [9, 10]. The heterodimer activates the transcription of several other clock genes, including those for period (PER) and cryptochrome (CRY) [11–13]. The resultant PER and CRY proteins heterodimerise, translocate to the nucleus, and inhibit the activity of CLOCK–BMAL1, thus forming a transcriptional–translational feedback loop. In parallel, the CLOCK–BMAL1 heterodimer activates the transcription of various clock-controlled genes [1, 2]. Given that some clock-controlled genes also serve as transcription factors, the expression of numerous genes may be tied to the functions of the circadian clock [1, 2]. Moreover, nearly half of the known nuclear receptors, including peroxisome proliferator-activated receptors (α, γ and δ) and thyroid hormone receptors (α and β), exhibit circadian expression in the liver and adipose tissues, providing an explanation for the cyclic behaviour of glucose and lipid metabolism [14].
Recently, the link between circadian clock function and metabolic diseases has attracted attention. Turek et al. [15] demonstrated that Clock mutant mice are hyperphagic and develop metabolic syndrome, hyperglycaemia and hyperlipidaemia. In addition, we showed that the rhythmic expression of clock genes is blunted in the liver and visceral adipose tissues of KK-Ay mice, a genetic model of type 2 diabetes [16]. In humans, genetic variations in the BMAL1 gene (also known as ARNTL) are reported to be associated with susceptibility to type 2 diabetes and hypertension [17]. Furthermore, CLOCK haplotypes are associated with metabolic syndrome [18] and non-alcoholic fatty liver disease [19]. These findings strongly indicate that dysfunction of the circadian clock contributes to the development of type 2 diabetes and metabolic syndrome. However, whether clock function is impaired in human patients with these metabolic diseases, as has been shown in mice, remains to be determined. To address this issue, we obtained peripheral leucocytes from patients with type 2 diabetes and from non-diabetic volunteers and compared their mRNA expression rhythms of clock genes.
Methods
Participants
Studies 1 and 2 were approved by the ethics committees of Kanazawa University (Kanazawa, Japan) and Medical Co. LTA (Fukuoka, Japan), respectively, and were conducted in accordance with the Declaration of Helsinki as revised in 2000. All individuals were Japanese and participated in the study after giving their written informed consent. We excluded the following individuals: those who had experienced either jet lag or shift work during the 2 weeks preceding the study, those who took psychotrophic drugs in the preceding month, and those with sleep disorder, inflammatory disease, malignancy or anaemia. Additional information about the lifestyles (habits, mealtimes and sleep time) was collected from all participants on the day of the study.
Study 1
The first study was performed from October to December 2006. Eight inpatients with type 2 diabetes and two non-diabetic inpatients with fatty liver were recruited from Kanazawa University Hospital (Kanazawa). All patients with diabetes met the American Diabetes Association’s diagnostic criteria for diabetes [20], whereas the other two were classified as having normal glucose tolerance and impaired fasting glucose, respectively, based on a 75 g OGTT. Four healthy men were also enrolled in this study. All of the 14 individuals kept regular hours for at least 2 weeks until the study day or hospital admission. Most individuals usually had three meals a day, whereas one healthy individual always skipped breakfast (ESM Fig. 2). As shown in Table 1, fasting glucose and HbA1c levels in patients with diabetes were significantly higher than those in non-diabetic individuals. In six of eight patients with diabetes, the disease was poorly controlled (HbA1c ≥ 7.0%). Three of the six patients were treated with oral agents (pioglitazone, glimepiride + metformin and glibenclamide + metformin + acarbose, respectively). Additionally, patients with diabetes were older than the non-diabetic individuals. The other variables did not differ between the groups.
On the day of the study, blood samples were taken from the forearm vein at 09:00, 15:00, 21:00 and 03:00 hours beginning at 09:00 hours. We chose these time points because BMAL1 and PER2 mRNA levels have been reported to peak at about 15:00 and 08:00 hours, respectively [21]. The healthy individuals were asked to assume their everyday routines and sleep in a dim room at their usual times. For the inpatients, sampling commenced within 48 h after admission and was conducted in their hospital room. Fasting blood samples for clinical chemistry were obtained from the inpatients in the early morning on the day after the admission and from the healthy individuals at least 2 weeks before the study day.
Study 2
The next study was carried out from November 2007 to January 2008. Twenty-six men with ages in their 50s to 60s were recruited from LTA PS Clinic (Fukuoka). Twelve individuals were outpatients with type 2 diabetes, whereas 14 individuals were healthy volunteers. All of the participants kept regular hours for at least 2 weeks until the study day. Most of them usually awoke between 05:00 and 07:00 hours and went to bed between 22:00 and 24:00 hours. One healthy participant consumed four meals a day, another healthy individual did not eat breakfast, and the other participants usually ate three meals a day. As shown in Table 2, seven patients were treated for type 2 diabetes with medications (glimepiride + metformin, n = 4; glimepiride + an α-glucosidase inhibitor, n = 2; glibenclamide + human insulin, n = 1). In addition, five and three patients were on medication to treat hypertension (an angiotensin II receptor antagonist, n = 4; amlodipine + olmesartan, n = 1) and hyperlipidaemia (pravastatin, n = 2; bezafibrate, n = 1), respectively. Venous blood samples for RNA isolation and blood chemistry were collected between 08:30 and 09:30 hours at the LTA PS Clinic after an overnight fast.
Isolation of leucocytes and purification of RNA
Immediately after blood sampling, leucocytes were isolated and stabilised using the LeukoLOCK Fractionation and Stabilization Kit (Applied Biosystems, Foster City, CA, USA). Briefly, 10 ml EDTA-anticoagulated blood was passed through a LeukoLOCK filter that captured only leucocytes, and the filter was flushed with PBS to remove residual erythrocytes. The filter was then filled with RNAlater to stabilise leucocyte RNA. The stabilised cells were stored on the filter at −20°C until RNA extraction. The isolation of total RNA was achieved using the LeukoLOCK Total RNA Isolation Kit (Applied Biosystems) according to the manufacturer’s instructions.
cDNA synthesis and real-time quantitative PCR
cDNA was synthesised from 1 μg total RNA using the high capacity cDNA reverse transcription kit (Applied Biosystems). Gene expression was analysed by real-time quantitative PCR using the Applied Biosystems 7900HT real-time PCR system, as previously described [16, 22]. All specific sets of primers and TaqMan probes (TaqMan gene expression assays) were obtained from Applied Biosystems. To control for variation in the amount of cDNA available for PCR in the different samples, gene expression levels of the target sequences were normalised to the expression of an endogenous control gene (GAPDH). The GenBank accession numbers, assay ID, and the target exons were NM_004898.2, Hs00231857_m1, 18-19 for CLOCK; NM_001178.4, Hs00154147_m1, 9-10 for BMAL1; NM_002616.1, Hs00242988_m1, 22-23 for PER1; NM_022817.1, Hs00256143_m1, 8-9 for PER2; NM_016831.1, Hs00213466_m1, 15-16 for PER3; NM_004075.2, Hs00172734_m1, 2-3 for CRY1; NM_001001928.2, Hs00947538_m1, 6-7 for PPARA; NM_138711.3, Hs01115512_m1, 4-5 for PPARG; NM_006238.3, Hs00602622_m1, 3-4 for PPARD; NM_002046.3, Hs99999905_m1, 3-3 for GAPDH; and NM_001002.3, Hs99999902_m1, 3-3 for ribosomal protein, large, P0 (RPLP0), respectively. Data were analysed using the comparative threshold cycle method.
Blood chemistry
Samples obtained after an overnight fast were assayed for plasma glucose, serum insulin, total cholesterol, HDL-cholesterol, triacylglycerols, aspartate aminotransferase, alanine aminotransferase and HbA1c. Each variable was measured using a commercial kit. Insulin sensitivity was estimated using the homeostasis model assessment for insulin resistance (HOMA-IR) [23].
Statistical analysis
Differences in the variables and mRNA levels between patients with diabetes and control individuals were evaluated using the Mann–Whitney test. The rhythmicity of each gene was assessed using the Friedman test. The values are presented as means ± SD, and p < 0.05 was considered significant. All calculations were performed using SPSS version 11 for Windows (SPSS Japan, Tokyo, Japan).
Results
Study 1
Because biological clock function in leucocytes is controversial [24], we first analysed the daily variation in mRNA expression of the clock genes in all 14 individuals. As shown in Fig. 1b–e, the mRNA expression of BMAL1, PER1, PER2 and PER3 exhibited slight but significant 24 h rhythmicity (χ 2 = 12.9, p < 0.01 for BMAL1; χ 2 = 22.9, p < 0.001 for PER1; χ 2 = 22.0, p < 0.001 for PER2; χ 2 = 25.0, p < 0.001 for PER3; Friedman test to evaluate rhythmicity). Similarly to previous reports [6–8, 21], the levels of PER1, PER2 and PER3 peaked in the early morning and dropped to a trough level in the evening. On the other hand, the mRNA levels of CLOCK, CRY1 (Fig. 1a,f) and another endogenous control gene (RPLP0) remained constant throughout the day (χ 2 = 6.8, p = 0.08 for CLOCK; χ 2 = 4.5, p = 0.21 for CRY1; χ 2 = 3.3, p = 0.34 for RPLP0; Friedman test). When the mRNA levels of clock genes were normalised to the expression of RPLP0, CRY1 (χ 2 = 11.4, p < 0.01) as well as BMAL1, PER1, PER2 and PER3 (χ 2 = 13.0–25.1, each p < 0.01) showed significant 24 h rhythms with a peak in the morning. These results support the idea that the circadian clock functions in leucocytes.
Daily mRNA expression profiles of clock genes in the peripheral leucocytes of patients with diabetes and non-diabetic individuals. a CLOCK; b BMAL1; c PER1; d PER2; e PER3; f CRY1; g PPARA; h PPARG; i PPARD. Peripheral leucocytes were obtained from six non-diabetic individuals (black circles) and eight patients with type 2 diabetes (white circles) at 09:00, 15:00, 21:00 and 03:00 hours. Transcript levels of the clock genes were determined by real-time quantitative PCR. The mean value of non-diabetic individuals at 09:00 hours was set to 1 for each gene. Means±SD. *p < 0.05, **p < 0.01 vs non-diabetic individuals
Next, we compared the mRNA expression rhythms of the clock genes in patients with diabetes with those in non-diabetic individuals (Fig. 1a–f). Interestingly, BMAL1 mRNA peaked in the evening in non-diabetic individuals (χ 2 = 8.0, p < 0.05; Friedman test to evaluate rhythmicity), whereas that peaked in the morning in patients with diabetes (χ 2 = 11.9, p < 0.01). In addition, the transcript levels of BMAL1, PER1, PER2 and PER3 were significantly lower in patients with diabetes than in non-diabetic individuals at one or more observation points. A multiple regression analysis identified that both PER1 and PER3 mRNA levels at 21:00 hours were correlated with the presence or absence of type 2 diabetes, but not with age (data not shown). Furthermore, the amplitudes of PER1 and PER3 were diminished in patients with diabetes (56% and 38% of those in non-diabetic patients; p = 0.06 and p < 0.01, respectively). The amplitude of PER3 determined using RPLP0 as an internal control also tended to be lower in patients with diabetes (72% of that in non-diabetic patients; p = 0.09). These results suggest that the function of the circadian clock was impaired in peripheral leucocytes of the patients with diabetes examined in this study.
Because peroxisome proliferator-activated receptors are known to exhibit circadian expression in liver and adipose tissues [14], their daily mRNA expression profiles in the leucocytes were determined as an indicator of the circadian clock function. The mRNA expression levels of PPARA and PPARD were maintained constant throughout the day (Fig. 1g–i; χ 2 = 5.5, p = 0.14 for PPARA; χ 2 = 4.7, p = 0.19 for PPARD), although those of patients with diabetes were significantly lower than those of non-diabetic individuals. On the other hand, PPARG mRNA showed a significant 24 h rhythm (χ 2 = 9.6, p < 0.05) with a peak in the small hours. This rhythmicity disappeared in patients with diabetes (χ 2 = 5.0, p = 0.18) as was expected. This result supports the view that the clock function was diminished in patients with diabetes.
Study 2
The patients with diabetes were older than the non-diabetic individuals in study 1. Because senescence might impair the circadian clock [25], the possibility exists that the age differences in part affected the results. Therefore, we next compared the transcript levels of the clock genes in patients with type 2 diabetes with those from age-matched healthy individuals. Moreover, we recruited only men for this study to exclude a sex effect suggested previously [16, 26]. As shown in Table 2, fasting glucose, HOMA-IR and HbA1c levels were significantly higher in patients with diabetes, but the other variables were similar between the two groups.
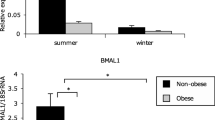
Consistent with the findings of study 1, the mRNA levels of PER1 at 09:00 hours were significantly lower in patients with diabetes than in control individuals (Fig. 2). Moreover, BMAL1 and PER3 mRNA levels were also lower in the patients. Conversely, no differences were observed between the two groups in the transcript levels of CLOCK, PER2, CRY1 or the endogenous control gene RPLP0. Note that BMAL1, PER1 and PER3 mRNA levels were inversely correlated with HbA1c levels (Spearman’s rank correlation coefficient ρ = −0.47, p < 0.05 for BMAL1; ρ = −0.52, p < 0.01 for PER1; ρ = −0.55, p < 0.01 for PER3; Fig. 3). Thus, this study provides evidence that mRNA expression of a subset of clock genes is diminished in patients with type 2 diabetes, especially those with poorly controlled blood glucose.
Transcript levels of the clock genes and RPLP0 in peripheral leucocytes of patients with diabetes and control individuals. Peripheral leucocytes were obtained from 14 healthy individuals (black circles) and 12 patients with type 2 diabetes (white circles) at 09:00 hours. Transcript levels of the target genes were determined by real-time quantitative PCR. The mean value of healthy individuals was set to 1 for each gene. *p < 0.05, **p < 0.01
Discussion
Recent studies have correlated metabolic diseases such as metabolic syndrome and type 2 diabetes with the circadian clock. Our previous study [16] revealed that the rhythmic expression of clock genes was slightly diminished in the peripheral tissues of mildly diabetic KK mice and was greatly blunted in severely diabetic KK-Ay mice. In this study, we demonstrated for the first time that the circadian clock of peripheral leucocytes is diminished in patients with type 2 diabetes, particularly in those with poorly controlled blood glucose.
High glucose has been shown to downregulate Per1 and Per2 mRNA expression in cultured fibroblasts [27]. In addition, Kohsaka et al. [26] reported that a high-fat diet affected the rhythmic mRNA expression of Clock, Bmal1 and Per2 in the liver and adipose tissues of mice. Considering these findings, alterations in glucose, lipid and energy metabolism and/or changes in the concentrations of humoral factors such as plasma glucose appear to influence the peripheral clock.
As shown in Fig. 1, human leucocytes exhibited very weak clock gene oscillations compared with the clocks of the liver and adipose tissues in mice [16]. The peripheral clocks are thought to be synchronised by the SCN through neural and humoral signals [2]. Because peripheral blood leucocytes, unlike the other peripheral tissues, are not controlled directly by neural signals, their oscillators might be easily desynchronised and greatly affected by humoral signals. The observation that the mRNA expression rhythms of PPARA and PPARD disappeared in leucocytes (Fig. 1g,i) supports this hypothesis. However, PPARG, as well as the clock genes, exhibited weak, but significant 24 h rhythmicity, suggesting that the each circadian clock in leucocytes works the same way as those in liver and adipose tissues. Whether each intracellular clock of individual leucocytes is impaired in patients with diabetes remains to be determined; however, attenuation of overall rhythmicity in peripheral leucocytes may cause leucocyte dysfunction. Leucocyte function is known to be depressed in patients with diabetes, which may contribute to their increased susceptibility to infection [28].
That the mutation and genotypes of core clock genes are associated with metabolic diseases [15, 17–19] leads us to speculate that the circadian clock contributes to the development of diabetes. Oishi et al. [29] demonstrated that clock function is preserved to a great extent in the livers, hearts and kidneys of mice with streptozotocin-induced insulinopenic diabetes. We could not confirm impairment of the circadian clock in the liver and adipose tissues of mice fed a high-fat diet, which was reported by Kohsaka et al. [26], even though the mice developed metabolic syndrome characterised by obesity, hyperlipidaemia and hyperglycaemia [30]. Thus, impairment of the circadian clock by type 2 diabetes or metabolic syndrome remains controversial. Further studies are needed to determine whether impaired clock function can be improved by glycaemic control in patients with type 2 diabetes.
Sedentary lifestyles and high dietary fat intake are thought to be instigators of metabolic diseases such as type 2 diabetes and metabolic syndrome. Additionally, it is not uncommon for modern people to keep irregular hours, live rather nocturnal lives or eat late-night snacks. Because light and dietary intake strongly entrain the master and peripheral clocks, respectively [2], these lifestyle features could cause malfunction of peripheral oscillators. Almost all of the individuals enrolled in this study kept regular hours and ordinary lifestyles. Nevertheless, their lives were rather nocturnal compared with the lives of ancient peoples. Therefore, the possibility exists that life in modern society could affect the biological clock, especially in highly susceptible individuals. Although the genotypes of the clock genes were not determined in this study, patients with type 2 diabetes might have defective or fragile circadian clocks.
In summary, rhythmic mRNA expression of clock genes was dampened in peripheral leucocytes of patients with type 2 diabetes. The impairment of the circadian clock appears to be closely associated with the pathophysiology of type 2 diabetes in humans.
Abbreviations
- BMAL1:
-
brain and muscle Arnt-like protein 1
- CLOCK:
-
clock homologue (mouse)
- HOMA-IR:
-
homeostasis model assessment for insulin resistance
- SCN:
-
suprachiasmatic nucleus
References
Lowrey PL, Takahashi JS (2004) Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet 5:407–441
Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418:935–941
Panda S, Antoch MP, Miller BH et al (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307–320
Storch KF, Lipan O, Leykin I et al (2002) Extensive and divergent circadian gene expression in liver and heart. Nature 417:78–83
Yoo SH, Yamazaki S, Lowrey PL et al (2004) PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101:5339–5346
Fukuya H, Emoto N, Nonaka H, Yagita K, Okamura H, Yokoyama M (2007) Circadian expression of clock genes in human peripheral leukocytes. Biochem Biophys Res Commun 354:924–928
Takimoto M, Hamada A, Tomoda A et al (2005) Daily expression of clock genes in whole blood cells in healthy subjects and a patient with circadian rhythm sleep disorder. Am J Physiol Regul Integr Comp Physiol 289:R1273–R1279
Kusanagi H, Mishima K, Satoh K, Echizenya M, Katoh T, Shimizu T (2004) Similar profiles in human period1 gene expression in peripheral mononuclear and polymorphonuclear cells. Neurosci Lett 365:124–127
Bunger MK, Wilsbacher LD, Moran SM et al (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009–1017
Gekakis N, Staknis D, Nguyen HB et al (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280:1564–1569
Kume K, Zylka MJ, Sriram S et al (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193–205
Okamura H, Miyake S, Sumi Y et al (1999) Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science 286:2531–2534
Vitaterna MH, Selby CP, Todo T et al (1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A 96:12114–12119
Yang X, Downes M, Yu RT et al (2006) Nuclear receptor expression links the circadian clock to metabolism. Cell 126:801–810
Turek FW, Joshu C, Kohsaka A et al (2005) Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308:1043–1045
Ando H, Yanagihara H, Hayashi Y et al (2005) Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology 146:5631–5636
Woon PY, Kaisaki PJ, Braganca J et al (2007) Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci U S A 104:14412–14417
Scott EM, Carter AM, Grant PJ (2008) Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes (Lond) 32:658–662
Sookoian S, Castano G, Gemma C, Gianotti TF, Pirola CJ (2007) Common genetic variations in CLOCK transcription factor are associated with nonalcoholic fatty liver disease. World J Gastroenterol 13:4242–4248
Genuth S, Alberti KG, Bennett et al (2003) Follow-up report on the diagnosis of diabetes mellitus. Diabetes Care 26:3160–3167
Archer SN, Viola AU, Kyriakopoulou V, von Schantz M, Dijk DJ (2008) Inter-individual differences in habitual sleep timing and entrained phase of endogenous circadian rhythms of BMAL1, PER2 and PER3 mRNA in human leukocytes. Sleep 31:608–617
Ando H, Oshima Y, Yanagihara H et al (2006) Profile of rhythmic gene expression in the livers of obese diabetic KK-A(y) mice. Biochem Biophys Res Commun 346:1297–1302
Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC (1985) Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28:412–419
Teboul M, Barrat-Petit MA, Li XM et al (2005) Atypical patterns of circadian clock gene expression in human peripheral blood mononuclear cells. J Mol Med 83:693–699
Kunieda T, Minamino T, Katsuno T et al (2006) Cellular senescence impairs circadian expression of clock genes in vitro and in vivo. Circ Res 98:532–539
Kohsaka A, Laposky AD, Ramsey KM et al (2007) High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6:414–421
Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y (2002) Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured rat-1 fibroblasts. J Biol Chem 277:44244–44251
Joshi N, Caputo GM, Weitekamp MR, Karchmer AW (1999) Infections in patients with diabetes mellitus. N Engl J Med 341:1906–1912
Oishi K, Kasamatsu M, Ishida N (2004) Gene- and tissue-specific alterations of circadian clock gene expression in streptozotocin-induced diabetic mice under restricted feeding. Biochem Biophys Res Commun 317:330–334
Yanagihara H, Ando H, Hayashi Y, Obi Y, Fujimura A (2006) High-fat feeding exerts minimal effects on rhythmic mRNA expression of clock genes in mouse peripheral tissues. Chronobiol Int 23:905–914
Acknowledgements
This study was supported by a Grant-in-Aid for Scientific Research (18790622 to H. Ando) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. We are grateful to all participants for their cooperation. We also thank M. Ishibashi, M. Nagamizu, T. Shobu and M. Kawagoe for their support.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material:
ESM Fig. 1
Diagram of the mammalian core circadian clock. (PDF 53 KB)
ESM Fig. 2
Representative lifestyle patterns of the participants. (PDF 11.9 KB)
Rights and permissions
About this article
Cite this article
Ando, H., Takamura, T., Matsuzawa-Nagata, N. et al. Clock gene expression in peripheral leucocytes of patients with type 2 diabetes. Diabetologia 52, 329–335 (2009). https://doi.org/10.1007/s00125-008-1194-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-008-1194-6