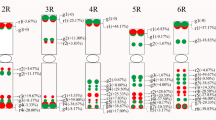
Abstract
PCR products from regions corresponding to sequences hybridising to wheat RFLP probes were sequenced in order to establish the level of DNA sequence variation among adapted wheat genotypes. Hexaploid bread wheat shows a very low rate of nucleotide polymorphism, approximately 1 polymorphic nucleotide per 1000 basepairs. Differences in PCR product length can be exploited to design genome-specific amplicons, which may have use in gene tagging or in diagnostic applications. Interpretation of results may be complicated by the simultaneous amplification of orthologous and paralogous sequences. These findings have significant implications for the use of STS markers in wheat and other polyploid species.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 3 October 1998 / Accepted: 28 November 1998
Rights and permissions
About this article
Cite this article
Bryan, G., Stephenson, P., Collins, A. et al. Low levels of DNA sequence variation among adapted genotypes of hexaploid wheat. Theor Appl Genet 99, 192–198 (1999). https://doi.org/10.1007/s001220051224
Issue Date:
DOI: https://doi.org/10.1007/s001220051224