Abstract
Key Message
The genetic architecture of RSA traits was dissected by GWAS and coexpression networks analysis in a maize association population.
Abstract
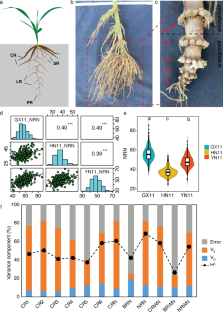
Root system architecture (RSA) is a crucial determinant of water and nutrient uptake efficiency in crops. However, the maize genetic architecture of RSA is still poorly understood due to the challenges in quantifying root traits and the lack of dense molecular markers. Here, an association mapping panel including 356 inbred lines were crossed with a common tester, Zheng58, and the test crosses were phenotyped for 12 RSA traits in three locations. We observed a 1.3 ~ sixfold phenotypic variation for measured RSA in the association panel. The association panel consisted of four subpopulations, non-stiff stalk (NSS) lines, stiff stalk (SS), tropical/subtropical (TST), and mixed. Zheng58 × TST has a 2.1% higher crown root number (CRN) and 8.6% less brace root number (BRN) than Zheng58 × NSS and Zheng58 × SS, respectively. Using a genome-wide association study (GWAS) with 1.25 million SNPs and correction for population structure, 191 significant SNPs were identified for root traits. Ninety (47%) of the significant SNPs showed positive allelic effects, and 101 (53%) showed negative effects. Each locus could explain 0.39% to 11.8% of phenotypic variation. By integrating GWAS results and comparing coexpression networks, 26 high-priority candidate genes were identified. Gene GRMZM2G377215, which belongs to the COBRA-like gene family, affected root growth and development. Gene GRMZM2G468657 encodes the aspartic proteinase nepenthesin-1, related to root development and N-deficient response. Collectively, our research provides progress in the genetic dissection of root system architecture. These findings present the further possibility for the genetic improvement of root traits in maize.






Similar content being viewed by others
Data availability
The genotype data for the association panel are available at http://www.maizego.org/Resources.html. All other data supporting the findings of this study are available within the paper, and its supplementary data is published online.
References
Andorf CM, Cannon EK, Portwood JL, Gardiner JM, Harper LC, Schaeffer ML, Braun BL, Campbell DA, Vinnakota AG, Sribalusu VV (2016) MaizeGDB update: new tools, data and interface for the maize model organism database. Nucleic Acids Res 44:D1195–D1201
Bates D, Mächler M, Bolker B, Walker S (2014) Fitting linear mixed-effects models using lme4. arXiv preprint arXiv:14065823
Bayuelo-Jiménez JS, Gallardo-Valdéz M, Pérez-Decelis VA, Magdaleno-Armas L, Ochoa I, Lynch JP (2011) Genotypic variation for root traits of maize (Zea mays L.) from the Purhepecha Plateau under contrasting phosphorus availability. Field Crop Res 121:350–362
Burton AL, Brown KM, Lynch JP (2013) Phenotypic diversity of root anatomical and architectural traits in Zea species. Crop Sci 53:1042–1055
Burton AL, Johnson JM, Foerster JM, Hirsch CN, Buell C, Hanlon MT, Kaeppler SM, Brown KM, Lynch JP (2014) QTL mapping and phenotypic variation for root architectural traits in maize (Zea mays L.). Theor Appl Genet 127:2293–2311
Burton AL, Johnson J, Foerster J, Hanlon MT, Kaeppler SM, Lynch JP, Brown KM (2015) QTL mapping and phenotypic variation of root anatomical traits in maize (Zea mays L.). Theor Appl Genet 128:93–106
Cai H, Chen F, Mi G, Zhang F, Maurer HP, Liu W, Reif JC, Yuan L (2012) Mapping QTLs for root system architecture of maize (Zea mays L.) in the field at different developmental stages. Theor Appl Genet 125:1313–1324
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 13:1194-1202
Chen Z, Sun J, Li D, Li P, He K, Ali F, Mi G, Chen F, Yuan L, Pan Q (2022) Plasticity of root anatomy during domestication of a maize-teosinte derived population. J Exp Bot 73:139–153
Das A, Schneider H, Burridge J, Ascanio AKM, Wojciechowski T, Topp CN, Lynch JP, Weitz JS, Bucksch A (2015) Digital imaging of root traits (DIRT): a high-throughput computing and collaboration platform for field-based root phenomics. Plant Methods 11:51
de Dorlodot S, Forster B, Pagès L, Price A, Tuberosa R, Draye X (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 12:474–481
Feldman L (1994) The maize root. The maize handbook. Springer, pp 29–37
Fu J, Cheng Y, Linghu J, Yang X, Kang L, Zhang Z, Zhang J, He C, Du X, Peng Z (2013) RNA sequencing reveals the complex regulatory network in the maize kernel. Nat Commun 4:1–12
Gu R, Chen F, Long L, Cai H, Liu Z, Yang J, Wang L, Li H, Li J, Liu W (2016) Enhancing phosphorus uptake efficiency through QTL-based selection for root system architecture in maize. J Genet Genomics 43:663–672
Guo J, Li C, Zhang X, Li Y, Zhang D, Shi Y, Song Y, Li Y, Yang D, Wang T (2020) Transcriptome and GWAS analyses reveal candidate gene for seminal root length of maize seedlings under drought stress. Plant Sci 292:110380
He X, Ma H, Zhao X, Nie S, Li Y, Zhang Z, Shen Y, Chen Q, Lu Y, Lan H (2016) Comparative RNA-Seq analysis reveals that regulatory network of maize root development controls the expression of genes in response to N stress. PLoS ONE 11:e0151697
Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, Peñagaricano F, Lindquist E, Pedraza MA, Barry K (2014) Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26:121–135
Hochholdinger F, Woll K, Sauer M, Dembinsky D (2004) Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programmes. Ann Bot 93:359–368
Hochholdinger F, Wen TJ, Zimmermann R, Chimot-Marolle P, Da Costa e Silva O, Bruce W, Lamkey KR, Wienand U, Schnable PS (2008) The maize (Zea mays L.) roothairless3 gene encodes a putative GPI-anchored, monocot-specific, COBRA-like protein that significantly affects grain yield. Plant J 54:888–898
Hoopes GM, Hamilton JP, Wood JC, Esteban E, Pasha A, Vaillancourt B, Provart NJ, Buell CR (2019) An updated gene atlas for maize reveals organ-specific and stress-induced genes. Plant J 97:1154–1167
Jin M, Zhang X, Zhao M, Deng M, Du Y, Zhou Y, Wang S, Tohge T, Fernie AR, Willmitzer L (2017) Integrated genomics-based mapping reveals the genetics underlying maize flavonoid biosynthesis. BMC Plant Biol 17:1–17
Kano M, Inukai Y, Kitano H, Yamauchi A (2011) Root plasticity as the key root trait for adaptation to various intensities of drought stress in rice. Plant Soil 342:117–128
Kassambara A, Mundt F (2017) Factoextra: extract and visualize the results of multivariate data analyses. R Package Version 1:337–354
Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21:2237–2252
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, Xiang Z, Song W, Ying K, Zhang M (2010) Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42:1027–1030
Li Q, Li L, Yang X, Warburton ML, Bai G, Dai J, Li J, Yan J (2010) Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2associated with kernel size and weight. BMC Plant Biol 10:1–15
Li Q, Yang X, Xu S, Cai Y, Zhang D, Han Y, Li L, Zhang Z, Gao S, Li J (2012) Genome-wide association studies identified three independent polymorphisms associated with α-tocopherol content in maize kernels. PLoS ONE 7:e36807
Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N (2013) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45:43–50
Li P, Chen F, Cai H, Liu J, Pan Q, Liu Z, Gu R, Mi G, Zhang F, Yuan L (2015) A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J Exp Bot 66:3175–3188
Li J, Chen F, Li Y, Li P, Wang Y, Mi G, Yuan L (2019) ZmRAP2.7, an AP2 transcription factor, is involved in maize brace roots development. Front Plant Sci 10:820
Li D, Wang H, Wang M, Li G, Chen Z, Leiser WL, Weiss TM, Lu X, Wang M, Chen S, Chen F, Yuan L, Wurschum T, Liu W (2021) Genetic dissection of phosphorus use efficiency in a maize association population under two P levels in the field. Int J Mol Sci 22(17):9311
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28:2397–2399
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12:e1005767
Liu Z, Gao K, Shan S, Gu R, Wang Z, Craft EJ, Mi G, Yuan L, Chen F (2017) Comparative analysis of root traits and the associated QTLs for maize seedlings grown in paper roll, hydroponics and vermiculite culture system. Front Plant Sci 8:436
Liu Z, Zhao Y, Guo S, Cheng S, Guan Y, Cai H, Mi G, Yuan L, Chen F (2019) Enhanced crown root number and length confers potential for yield improvement and fertilizer reduction in nitrogen-efficient maize cultivars. Field Crop Res 241:107562
Liu S, Barrow CS, Hanlon M, Lynch JP, Bucksch A (2021) DIRT/3D: 3D root phenotyping for field-grown maize (Zea mays). Plant Physiol 187:739–757
Lynch J (1995) Root architecture and plant productivity. Plant Physiol 109:7
Lynch JP (2018) Rightsizing root phenotypes for drought resistance. J Exp Bot 69:3279–3292
Ma L, Qing C, Frei U, Shen Y, Lübberstedt T (2020) Association mapping for root system architecture traits under two nitrogen conditions in germplasm enhancement of maize doubled haploid lines. The Crop Journal 8:213–226
Mi G, Chen F, Yuan L, Zhang F (2016) Ideotype root system architecture for maize to achieve high yield and resource use efficiency in intensive cropping systems. Adv Agron 139:73–97
Muszynski MG, Moss-Taylor L, Chudalayandi S, Cahill J, Del Valle-Echevarria AR, Alvarez-Castro I, Petefish A, Sakakibara H, Krivosheev DM, Lomin SN (2020) The maize hairy sheath frayed1 (Hsf1) mutation alters leaf patterning through increased cytokinin signaling. Plant Cell 32:1501–1518
Pace J, Gardner C, Romay C, Ganapathysubramanian B, Lübberstedt T (2015) Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC Genomics 16:1–12
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Human Genet 81:559–575
Raihan MS, Liu J, Huang J, Guo H, Pan Q, Yan J (2016) Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58 × SK maize population. Theor Appl Genet 129:1465–1477
Ren W, Zhao L, Liang J, Wang L, Chen L, Li P, Liu Z, Li X, Zhang Z, Li J, He K, Zhao Z, Ali F, Mi G, Yan J, Zhang F, Chen F, Yuan L, Pan Q (2022) Genome-wide dissection of changes in maize root system architecture during modern breeding. Nat Plants 8(12):1408–1422
Saengwilai P, Tian X, Lynch JP (2014) Low crown root number enhances nitrogen acquisition from low-nitrogen soils in maize. Plant Physiol 166:581–589
Schaefer RJ, Michno J-M, Jeffers J, Hoekenga O, Dilkes B, Baxter I, Myers CL (2018) Integrating coexpression networks with GWAS to prioritize causal genes in maize. Plant Cell 30:2922–2942
Shin J-H, Blay S, Lewin-Koh N, McNeney B, Yang G, Reyers M, Yan Y, Graham J (2016) Package ‘LDheatmap’. R package
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432
Steklov MY, Lomin SN, Osolodkin DI, Romanov GA (2013) Structural basis for cytokinin receptor signaling: an evolutionary approach. Plant Cell Rep 32:781–793
Stelpflug SC, Sekhon RS, Vaillancourt B, Hirsch CN, Buell CR, de Leon N, Kaeppler SM (2016) An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9(1):plantgenome2015–plantgenome2104
Sun B, Gao Y, Lynch JP (2018) Large crown root number improves topsoil foraging and phosphorus acquisition. Plant Physiol 177:90–104
Team RC (2013) R: A language and environment for statistical computing
Thorup-Kristensen K, Halberg N, Nicolaisen M, Olesen JE, Crews TE, Hinsinger P, Kirkegaard J, Pierret A, Dresbøll DB (2020) Digging deeper for agricultural resources, the value of deep rooting. Trends Plant Sci 25:406–417
Trachsel S, Kaeppler SM, Brown KM, Lynch JP (2011) Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341:75–87
Tracy SR, Nagel KA, Postma JA, Fassbender H, Wasson A, Watt M (2020) Crop improvement from phenotyping roots: highlights reveal expanding opportunities. Trends Plant Sci 25:105–118
Viana WG, Scharwies JD, Dinneny JR (2022) Deconstructing the root system of grasses through an exploration of development, anatomy and function. Plant, Cell Environ 45:602–619
Walbot V (2009) 10 reasons to be tantalized by the B73 maize genome. PLoS Genet 5:e1000723–e1000723
Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW (1997) The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev 11:799–811
Wang H, Xu C, Liu X, Guo Z, Xu X, Wang S, Xie C, Li W-X, Zou C, Xu Y (2017) Development of a multiple-hybrid population for genome-wide association studies: theoretical consideration and genetic mapping of flowering traits in maize. Sci Rep 7:40239
Wang K, Zhang Z, Sha X, Yu P, Li Y, Zhang D, Liu X, He G, Li Y, Wang T, Guo J, Chen J, Li C (2023) Identification of a new QTL underlying seminal root number in a maize-teosinte population. Front Plant Sci 14:1132017
Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, Yan J (2014) Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun 5:3438
Wickham H (2016) ggplot2: elegant graphics for data analysis Springer-Verlag New York; 2009. Book
Wu B, Ren W, Zhao L, Li Q, Sun J, Chen F, Pan Q (2022) Genome-wide association study of root system architecture in maize. Genes (basel) 13(2):181
Xue X-H, Guo C-Q, Du F, Lu Q-L, Zhang C-M, Ren H-Y (2011) AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Mol Plant 4:264–278
Yan J, Kandianis CB, Harjes CE, Bai L, Kim E-H, Yang X, Skinner DJ, Fu Z, Mitchell S, Li Q, Fernandez MGS, Zaharieva M, Babu R, Fu Y, Palacios N, Li J, DellaPenna D, Brutnell T, Buckler ES, Warburton ML, Rocheford T (2010) Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat Genet 42:322–327
Yan J, Warburton M, Crouch J (2011) Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Sci 51:433–449
Yang X, Gao S, Xu S, Zhang Z, Prasanna BM, Li L, Li J, Yan J (2011) Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol Breeding 28:511–526
Yin L (2020) CMplot: circle manhattan plot. R Package Version 3:6
Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H (2004) Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiol 134:1654–1661
Yu P, Hochholdinger F, Li C (2015) Root-type-specific plasticity in response to localized high nitrate supply in maize (Zea mays). Ann Bot 116:751–762
Yu T, Liu C, Lu X, Bai Y, Zhou L, Cai Y (2019) ZmAPRG, an uncharacterized gene, enhances acid phosphatase activity and Pi concentration in maize leaf during phosphate starvation. Theor Appl Genet 132:1035–1048
Zhang X, Warburton ML, Setter T, Liu H, Xue Y, Yang N, Yan J, Xiao Y (2016) Genome-wide association studies of drought-related metabolic changes in maize using an enlarged SNP panel. Theor Appl Genet 129:1449–1463
Zheng Z, Hey S, Jubery T, Liu H, Yang Y, Coffey L, Miao C, Sigmon B, Schnable JC, Hochholdinger F, Ganapathysubramanian B, Schnable PS (2019) Shared Genetic Control of Root System Architecture between Zea mays and Sorghum bicolor 1 [OPEN]. Plant Physiol 182:977–991
Zhu J, Kaeppler SM, Lynch JP (2005) Mapping of QTL controlling root hair length in maize (Zea mays L.) under phosphorus deficiency. Plant Soil 270:299–310
Acknowledgements
The authors gratefully acknowledge Dr. Jianbin Yan, Huazhong Agricultural University, who provided the germplasm resources and established the genotypes for the association mapping population.We also thank Dr. Philip James Kear, from International Potato Center–China Center for Asia and the Pacific, for providing valuable feedback and editing the revised version of our manuscript.
Funding
This study was financially supported by the Hainan Provincial Natural Science Foundation of China (321CXTD443) and the National Natural Science Foundation of China (31972485, 31971948).
Author information
Authors and Affiliations
Contributions
FC and QP designed the experiment; ZL analyzed the data and wrote the manuscript; PL performed the experiments; WR and ZC assisted in data analysis; and OT, GM, LY, FC, and QP contributed to manuscript editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2023_4442_MOESM1_ESM.xlsx
Supplement Table S1: Information of the 356 representative maize panel. Supplement Table S2: Environmental information of Guangxing, Yunnan and Hainan locations for field experiments in this study. Supplement Table S3: Summary of the significant SNPs association loci for root traits. (XLSX 43 KB)
122_2023_4442_MOESM2_ESM.tif
Supplement Figure 1 Distribution of 1.25 million polymorphic SNPs in the maize genome. Heatmap of SNP density on the chromosome within a 1-Mb interval, colors were used to indicate the number of SNPs within the 1-Mb interval. The physical position of the SNPs was based on the B73 reference sequence (RefGen_V2). (TIF 9090 KB)
122_2023_4442_MOESM3_ESM.tif
Supplement Figure 2 Distribution and correlation of root system architecture traits between each pair of locations. The significance levels of pairwise t-tests were added. * indicates P ≤ 0.05, ** indicates P ≤ 0.01; *** indicates P ≤ 0.001; **** indicates P ≤ 0.0001. Different lowercase letters indicate significant differences (P < 0.05) in different locations, as determined by Tukey's HSD test. Abbreviations for root traits are as follows: BRN, brace root number; BRWN, brace root whorl number; CR1, 1st whorl crown roots; CR2, 2nd whorl crown roots; CR3, 3rd whorl crown roots; CR4, 4th whorl crown roots; CR5, 5th whorl crown roots; CR6, 6th-8th whorl crown roots; CRN, crown root number; CRWN, crown root whorl number; NRWN, nodal root whorl number. GX11: Guangxi location in 2011, HN11: Hainan location in 2011, YN11: Yunnan location in 2011. (TIF 9318 KB)
122_2023_4442_MOESM4_ESM.tif
Supplement Figure 3 The phenotypic distribution of root traits in the association panel. Abbreviations for root traits are as follows: BRN, brace root number; BRWN, brace root whorl number; CR1, 1st whorl crown roots; CR2, 2nd whorl crown roots; CR3, 3rd whorl crown roots; CR4, 4th whorl crown roots; CR5, 5th whorl crown roots; CR6, 6th-8th whorl crown roots; CRN, crown root number; CRWN, crown root whorl number; NRWN, nodal root whorl number. (TIF 65415 KB)
122_2023_4442_MOESM5_ESM.tif
Supplement Figure 4 Phenotypic correlations among the root traits in the association panel. Abbreviations for root traits are as follows: BRN, brace root number; BRWN, brace root whorl number; CR1, 1st whorl crown roots; CR2, 2nd whorl crown roots; CR3, 3rd whorl crown roots; CR4, 4th whorl crown roots; CR5, 5th whorl crown roots; CR6, 6th-8th whorl crown roots; CRN, crown root number; CRWN, crown root whorl number; NRWN, nodal root whorl number. (TIF 61613 KB)
122_2023_4442_MOESM6_ESM.tif
Supplement Figure 5 The percentage of total variance explained by each principal component. Abbreviations for root traits are as follows: BRN, brace root number; BRWN, brace root whorl number; CR1, 1st whorl crown roots; CR2, 2nd whorl crown roots; CR3, 3rd whorl crown roots; CR4, 4th whorl crown roots; CR5, 5th whorl crown roots; CR6, 6th-8th whorl crown roots; CRN, crown root number; CRWN, crown root whorl number; NRN, nodal root number; NRWN, nodal root whorl number. (TIF 11460 KB)
122_2023_4442_MOESM7_ESM.tif
Supplement Figure 6 Comparison of root traits between PA and SPT heterotic groups. Different lowercase letters indicate significant differences (P < 0.05) in different locations, as determined by Tukey's HSD test. Abbreviations for root traits are as follows: BRN, brace root number; BRWN, brace root whorl number; CR1, 1st whorl crown roots; CR2, 2nd whorl crown roots; CR3, 3rd whorl crown roots; CR4, 4th whorl crown roots; CR5, 5th whorl crown roots; CR6, 6th-8th whorl crown roots; CRN, crown root number; CRWN, crown root whorl number; NRN, nodal root number; NRWN, nodal root whorl number. (TIF 11656 KB)
122_2023_4442_MOESM8_ESM.tif
Supplement Figure 7 Quantile–quantile plots of root traits. (a) BRN, brace root number, (b) BRWN, brace root whorl number, (c) CR1, 1st whorl crown roots, (d) CR2, 2nd whorl crown roots, (e) CR3, 3rd whorl crown roots, (f) CR4, 4th whorl crown roots, (g) CR5, 5th whorl crown roots, (h) CR6, 6th-8th whorl crown roots, (i) CRN, crown root number, (j) CRWN, crown root whorl number, (k) NRN, nodal root number, (l) NRWN, nodal root whorl number. (TIF 60252 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Z., Li, P., Ren, W. et al. Hybrid performance evaluation and genome-wide association analysis of root system architecture in a maize association population. Theor Appl Genet 136, 194 (2023). https://doi.org/10.1007/s00122-023-04442-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04442-7