Abstract
Key message
Identification and functional analysis of the male sterile gene MS6 in Glycine max.
Abstract
Soybean (Glycine max (L.) Merr.) is an important crop providing vegetable oil and protein. The male sterility-based hybrid breeding is a promising method for improving soybean yield to meet the globally growing demand. In this research, we identified a soybean genic male sterile locus, MS6, by combining the bulked segregant analysis sequencing method and the map-based cloning technology. MS6, highly expressed in anther, encodes an R2R3 MYB transcription factor (GmTDF1-1) that is homologous to Tapetal Development and Function 1, a key factor for anther development in Arabidopsis and rice. In male sterile ms6 (Ames1), the mutant allele contains a missense mutation, leading to the 76th leucine substituted by histidine in the DNA binding domain of GmTDF1-1. The expression of soybean MS6 under the control of the AtTDF1 promoter could rescue the male sterility of attdf1 but ms6 could not. Additionally, ms6 overexpression in wild-type Arabidopsis did not affect anther development. These results evidence that GmTDF1-1 is a functional TDF1 homolog and L76H disrupts its function. Notably, GmTDF1-1 shows 92% sequence identity with another soybean protein termed as GmTDF1-2, whose active expression also restored the fertility of attdf1. However, GmTDF1-2 is constitutively expressed at a very low level in soybean, and therefore, not able to compensate for the MS6 deficiency. Analysis of the TDF1-involved anther development regulatory pathway showed that expressions of the genes downstream of TDF1 are significantly suppressed in ms6, unveiling that GmTDF1-1 is a core transcription factor regulating soybean anther development.
Similar content being viewed by others
Introduction
Soybean (Glycine max (L.) Merr.) is a major crop providing plant protein and oil in the food supply, but it has a relatively low yield (Palmer et al. 2001). Hybrid breeding technology, which significantly increases the yield of major crops such as rice and maize, shows a great potential in improving the yield of soybean as well (Kim and Zhang 2018; Palmer et al. 2001). For instance, HybSoy1, the first officially approved and commercially applicable hybrid soybean, can increase the yield by 20.8% (Zhao et al. 2004). In the hybrid breeding system, male sterile lines are indispensable for avoiding the time-consuming and tedious artificial emasculation.
There are two types of male sterility (MS), i.e., genic male sterility (GMS) controlled by nuclear MS genes and cytoplasmic male sterility (CMS) co-conditioned by mitochondrial CMS genes and nuclear fertility restorer genes (Guo and Liu 2012). Based on the stability of the MS phenotype, GMS could be further subdivided into two groups, the stable GMS and the environmental-sensitive GMS (EGMS). The hybrid breeding systems applied today include the CMS-based three-line system and the EGMS-based two-line system (Kim and Zhang 2018). However, for the CMS-based three-line system, the genetic resources of fertility restorer lines are limited. Some types of CMS cytoplasm even show detrimental effects on crop performance, such as elevating disease susceptibility in maize T-type CMS lines (Levings 1990). As to the EGMS two-line system, the sterility of EGMS lines could be reversed when the growth environment changes unexpectedly, endangering the hybrid production process (Chen et al. 2011a). Comparatively, stable GMS lines can overcome the limitations of EGMS and CMS lines due to the stable sterility and wide-range choices of restorer lines. However, lack of maintainer lines has been restricted the application of GMS lines for a long time. Until recently, the transgenic-based Seed Production Technology (SPT) system, which creates artificial maintainer lines by introducing a fertility restorer gene, a pollen killer gene and a selection marker gene into the GMS lines, makes GMS applicable to hybrid production (Perez-Prat and van Lookeren Campagne 2002; Weber et al. 2009).
So far, 13 GMS loci have been reported in soybean, which are distributed on seven chromosomes and function in conditioning anther development, including ms1-ms9, msp, msMOS, mst-M and msNJ (Yang et al. 2014; Zhao et al. 2019; Nie et al. 2019; Thu et al. 2019). Deficiency of these loci confers recessive sporophytic male-sterile phenotypes. Among these mutants, ms6 displays a stable non-pollen phenotype (Skorupska and Palmer 1989; Ilarslan et al. 1999), making it an idea material for developing the SPT system in soybean. There are two independent, spontaneous ms6 mutants maintained as heterozygotes in Soybean Genetic Type Collection as T295H (+ /ms6 (Ames1)) (Skorupska and Palmer 1989) and T354H (+ /ms6 (Ames2)) (Ilarslan et al. 1999), respectively. Comparative microscopic studies on fertile and sterile offspring from T354H showed that the cytological abnormalities in ms6 anther firstly appear at the microspore mother cell (MMC) stage on tapetal and parietal layers, which possess more vacuoles in cells. Then, the tapetum in ms6 undergoes a premature programmed cell death and degrades entirely in the late microspore stage, while the parietal layer in ms6 is enlarged during the later development stages (Ilarslan et al. 1999). The reproductive cells in ms6 exhibit abnormalities at the end of meiosis, forming partially separated tetrads, which subsequently collapse in the late microspore stage when the fertile microspores are processing the first mitosis (Ilarslan et al. 1999). The similar phenomenon was observed during the microsporogenesis in ms6 (Ames1) mutant from T295H (Skorupska and Palmer 1989).
Previous studies have revealed that the ms6 mutation is closely linked to the flower-color gene W1 (Skorupska and Palmer 1989; Palmer et al. 1998; Ilarslan et al. 1999), and located in a 3.7 Mb region on chromosome 13 (Chr13) between simple sequence repeat (SSR) markers Satt030 and Satt149 (Yang et al. 2014). In this study, via BSA-sequencing and map-based cloning experiments, we further narrowed down the genetic region of ms6 (Ames1) and identified the corresponding mutation, which is a missense mutation in the gene Glyma.13G066600 (designated as MS6). MS6 encodes a homolog of Tapetal Development and Function 1 (TDF1), an R2R3 MYB transcription factor, which is critical for regulating tapetal layer degeneration in Arabidopsis and rice (Zhu et al. 2008; Cai et al. 2015). Accordingly, we named the protein encoded by MS6 as GmTDF1-1. Notably, all the components constituting the TDF1-involved regulatory pathway in Arabidopsis and rice were also found in soybean, but each has multiple paralogs, in which the expressions of genes acting downstream of TDF1 were strongly suppressed in ms6 flowers. These indicate that the TDF1-related regulatory pathway is likely conserved but more complicated in soybean due to the recent whole-genome duplication events. Results from this study provide new insights into the regulatory network in soybean anther development, and make the ms6 mutant a practicable material for the SPT system to facilitate hybrid seed production in soybean.
Materials and methods
Primers
Primers used in the present study were listed in Table S1.
Plant materials and growth conditions
The ms6 mutant used in this study was derived from T295H (PI 533601, + /ms6 (Ames1)), which was achieved from the collection of the National Plant Germplasm System (NPGS) in the United States. Allele ms6 (Ames1) is referred to as ms6 hereafter. To narrow down the genetic region of the ms6 mutation, a BC5F2 segregating population was developed by using T295H as ms6 donor and a wild-type (WT) cultivar ‘JiuB’, from Jilin, China, as a recurrent male parent. Additionally, 40 male-fertile accessions with different geographic origins, including 21 improved cultivars, 4 landraces and 15 wild soybeans (G. soja), were used for evaluating the conservation of the nucleotide mutated in the ms6 allele (Table S2). The mapping population was planted on the farm of Fanjiatun, Jilin in summer. For other studies, soybean plants were grown in pots (two plants per pot) outdoors in summer and in the greenhouse in winter at 28 °C with a photoperiod of 16 h light /8 h dark, in Xi’an, China.
Arabidopsis thaliana and Nicotiana benthamiana plants were grown in soil in the greenhouse at 22 °C with a photoperiod of 16 h light /8 h dark, in Xi’an, China. The Arabidopsis germplasms used in this study were WT Columbia (Col), heterozygous attdf1 mutant (+ /attdf1) in Col background (obtained from Dr. Zhongnan Yang’s lab) and transgenic lines for the complementary and overexpression experiments. The attdf1 mutation is caused by a single nucleotide polymorphism (SNP) that leads to a premature stop codon in AT3G28470 (AtTDF1) and simultaneously creates an MseI site (Zhu et al. 2008).
Morphological and cytological analyses
For general morphological observation of ms6 anthers, soybean flowers one day before blooming were collected from the fertile and sterile descendants of T295H (+ /ms6). Stamens were dissected and imaged under a stereo microscope Nikon SMZ25. Pollens were squeezed out, stained with 1% I2-KI solution and photographed under a light microscope Leica DM2500. For Arabidopsis, mature anthers before anthesis were collected from WT, homozygous attdf1 mutant and various transgenic lines, stained with Alexander staining buffer (Peterson et al. 2010) and photographed under Leica DM2500.
For cytological analysis, flowers at the late tetrad and early pollen stages were collected from the fertile and sterile descendants of T295H (+ /ms6) and immediately immersed into the FAA fixation solution. After dehydration, flower samples were imbedded into resin with Technovit H7100-GMA kit (Heraeus Kulzer, Germany) following the manufacturer’s instruction and sliced into 2-μm transverse sections with Leica RM2265. Sections on slides were stained with 0.5% toluidine blue staining buffer and imaged under Leica DM2500 after sealed.
DNA extraction, BSA-sequencing (BSA-seq) and fine mapping analyses
Genomic DNA samples were extracted from young leaves with the Nuclean Plant Genomic DNA Kit (CWBIO, China) for regular PCR analysis and BSA-seq experiment. For BSA-seq analysis, two bulks were constructed from the BC5F2 mapping population. One was composed of 20 homozygous WT plants and the other was composed of 20 homozygous ms6 plants. Genomic DNA isolated from each bulk was fractioned to build a 350-bp pair-end (PE) sequencing library and sequenced on Illumina NovaSeq 6000 PE150 platform in Novogene Company (China). SNPs and InDels (insertions-deletions) of each bulk were annotated using the Wm82.a2.v1 genome as the reference. The SNP-index of each bulk and the difference between the SNP-index of two bulks, ∆(SNP-index), were calculated as previously described (Takagi et al. 2013). Then, ∆(SNP-index) values were plotted against their genome positions to identify the region associated with ms6.
Polymorphic SSR markers in the genetic window of ms6 identified by BSA-seq were further used to screen the ms6 individuals in the BC5F2 population via canonical PCR. The fragments amplified with SSR primers were resolved on 8% polyacrylamide gel in 1xTAE buffer by electrophoresis and visualized by the silver staining method (Bassam et al. 1991). The genetic map was constructed from the data with MAPMAKER 3.0 (Lander et al. 1987).
Cleaved amplified polymorphic sequence (CAPS) analysis
CAPS analysis was performed to genotype the MS6 locus in soybean and the AtTDF1 locus in Arabidopsis. The genomic regions of MS6 or AtTDF1 spanning the SNP site were amplified from testing samples by PCR with gene specific primers. Subsequently, five microliters of PCR products were digested with MseI according to the manufacturer’s instruction, and the band patterns were analyzed on 4% agarose gel.
Bioinformatics and phylogenetic analyses
Conserved structural domain in the GmTDF1-1 polypeptide was predicted by the simple modular architecture research tool (SMART) (http://smart.embl-heidelberg.de/), showing GmTDF1-1 contained a typical R2R3 MYB DNA-binding domain. The amino acid sequences of the R2 region of GmTDF1-1 and 23 well-characterized MYB proteins in Arabidopsis (Dubos et al. 2010) were aligned with Bioedit software (Tom Hall, Ibis Biosciences, U.S.) to access the conservation of the R2 region of GmTDF1-1. The proteins with highest homology to GmTDF1-1 in G. max (GmTDF1-2), A. thaliana (AtTDF1) and Oryza sativa (OsTDF1) were identified in NCBI Database with BLASTP, and the conservation between these three proteins and GmTDF1-1 was evaluated with Bioedit software as well.
The amino acid sequences of 17 proteins were subjected to phylogenetic analysis, including GmTDF1-1, GmTDF1-2, AtTDF1, OsTDF1 and the putative TDF1 orthologs in 13 additional representative species from different land plant evolutionary lineages, including three dicot (Medicago truncatula, Vitis vinifera and Solanum lycopersicum), five monocot (Zea mays, Sorghum bicolor, Ananas comosus, Musa acuminate and Zostera marina), one basal angiosperm (Amborella trichopoda), two gymnosperm (Ginkgo biloba and Pinus taeda), one lycophyte (Selaginella moellendorffii) and one moss (Physcomitrella patens) species. The sequences of the putative TDF1 orthologs were retrieved from NCBI with BLASTP by using AtTDF1 (NP 189,488.1) and OsTDF1 (XP 015,630,216.1) as query peptides, and the one with the highest bit-score in each species was selected. AtMYB80 (NP 200,422.1) and OsMYB80 (XP 015,635,420.1) were used as outgroup sequences. All the protein sequences were aligned using the ClustalW2 with default parameters, and the phylogenetic tree was constructed by the neighbor-joining method with 1000 bootstrap replicates using MEGA 6 (Tamura et al. 2013).
RNA extraction, RT-PCR and qRT-PCR analyses
DNA-free total RNA was extracted from desired tissues with RNAprep Pure Plant Kit (Tiangen, China). One microgram of RNA was reverse transcribed to cDNA using PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Japan). RT-PCR was conducted with rTaq DNA polymerase (Takara, Japan) for analyzing gene expression or with 2 × PrimeSTAR Max Premix (Takara, Japan) for cloning purposes following the manufacturer’s instructions. For quantitative RT-PCR, one microliter of 1:10 diluted cDNA sample was used as a template, and the qRT-PCR reaction was conducted as previously described (Zhang et al. 2018a). The relative expression levels of MS6 and GmTDF1-2 in various WT tissues were calculated with the 2−∆Ct method (Livak and Schmittgen 2001). The fold-changes of target genes in young flowers (2.5–3.5 mm in length) of ms6 compared to those of WT were calculated with the 2−∆∆Ct method (Livak and Schmittgen 2001). All data were normalized against the expression level of GmActin11 (Glyma.18G290800). Three biological replicates, each with three-technique replicates, were performed for each sample.
Subcellular localization analysis
The coding sequences (CDS) for proteins GmTDF1-1 and GmTDF1-1L76H (the mutant protein encoded by the ms6 allele) without stop codon were amplified by RT-PCR from WT and ms6 flowers, respectively, and cloned into the XbaI site upstream of the GFP gene in the binary vector pLM-35S-GFP to create pLM-35S-GmTDF1-1-GFP and pLM-35S-GmTDF1-1L76H-GFP, respectively. Vectors were transformed into Agrobacterium tumefaciens strain GV3101 and infiltrated into 4-week-old N. benthamiana leaves. GFP signals in leaves were observed and imaged at 48 h post infiltration under an Olympus Fluoview FV1000 confocal laser scanning microscope.
Transactivation activity assay in yeast
The CDSs for GmTDF1-1 and its DNA binding domain (amino acids 1–191, GmTDF1-1DBD) were amplified by RT-PCR from WT flowers; the CDS for GmTDF1-1L76H was amplified by RT-PCR from ms6 flowers. The obtained fragments were cloned into the pGBKT7 vectors downstream of the CDS for GAL4-BD, respectively, to generate the vectors of pGBKT7-GmTDF1-1, pGBKT7-GmTDF1-1DBD and pGBKT7-GmTDF1-1L76H, which were subsequently transformed into Saccharomyces cerevisiae strain AH109 via one-step transformation method (Chen et al. 1992). After selected on the synthetic dropout medium lack of trypsin (SD/-Trp), the positive colonies were diluted into the same concentration with autoclaved ddH2O. Then, five microliters of cell suspensions were dropped on the selective medium SD/-Trp/-His and grown for 3–4 days under 30 °C to evaluate the activation activities of target proteins.
Complementary and overexpression analyses
For complementary analysis, the 817-bp AtTDF1 promoter reported previously by Zhu et al. (2008) was cloned into pCAMBIA1301 via KpnI and XbaI to get the pCAMBIA1301-AtTDF1p vector. The CDSs of MS6, ms6, AtTDF1, AtTDF1L46H and GmTDF1-2 were cloned into pCAMBIA1301-AtTDF1p downstream the AtTDF1 promoter through XbaI and BstEII to acquire pCAMBIA1301-AtTDF1p-MS6, pCAMBIA1301-AtTDF1p-ms6, pCAMBIA1301-AtTDF1p-AtTDF1, pCAMBIA1301-AtTDF1p-AtTDF1L46H and pCAMBIA1301-AtTDF1p-GmTDF1-2, respectively. Here, AtTDF1L46H is a variant of AtTDF1 CDS generated by site-directed mutagenesis via overlap extension PCR (Ho et al. 1989), and it encodes a mutant form of AtTDF1 (AtTDF1L46H) corresponding to GmTDF1-1L76H.
A. tumefaciens strain GV3101 carrying these vectors were used to transform Arabidopsis attdf1 heterozygous plants by the floral-dip method (Clough and Bent 1998). T1 transformants were screened on 1/2 MS agar plates supplemented with 1% sucrose and 20 mg/L hygromycin. The hygromycin-resistant T1 plants were transplanted into soil for further growth after they were verified for the presence of the transgene by PCR. The native AtTDF1 locus in each T1 transgenic plant was genotyped by CAPS analysis. Transgenic plants in homozygous attdf1 background were further scored for fertility.
For overexpression analysis, the CDS of the ms6 allele was cloned into the PLM-35S vector via XbaI and EcoRI site to generate PLM-35S-ms6, which was transformed into WT Arabidopsis (Col) via Agrobacterium as described above. T1 transformants were selected by Basta resistance, confirmed by PCR analysis and further scored for anther fertility. The expression level of ms6 in T1 transgenic plants was examined by RT-PCR with AtActin as a reference gene.
Results
Phenotypic characterization of the ms6 mutant
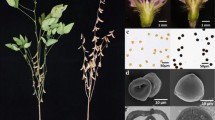
The offsprings of heterozygous ms6 plants (T295H) were planted in the greenhouse. During the vegetative stage, all plants grew well just like WT plants, while at the reproductive stage about a quarter of plants were male-sterile and failed to develop pods, indicating they were ms6 homozygotes (Fig. 1a). The anthers of ms6 plants were whitish and shrunken compared to those of WT plants (Fig. 1b). Moreover, pollen grains released from WT anthers were round and stained dark blue by I2-KI solution (Fig. 1c), but no pollen grains were produced in ms6 anthers (Fig. 1d). These results are consistent with the previous report (Skorupska and Palmer 1989).
Phenotypic characterization of ms6 in soybean a Wild-type (WT) and ms6 plants at R3 stage of WT. Compared with WT, ms6 fails to produce elongated pods at nodes (red arrow). Scale bar = 10 cm. b WT and ms6 flowers with petals and sepals removed. Scale bar = 0.5 mm. c, d I2-KI staining of pollens squeezed out from WT and ms6 anthers. Scale bar = 50 μm. e–h Semi-thin sections of WT and ms6 anther lobes at the late tetrad stage e, f and the early pollen stage g, h. Scale bar = 50 μm. Red triangles indicate the multi-nuclei in tetrads. E, Epidermis; En, Endodermis; M, Middle layer; P, Parietal layer; T, Tapetal layer; Tds, Tetrads; Pol, Pollen (color figure online)
The flowers of WT and ms6 at the late tetrad and early pollen stages were cross-sectioned and observed under light microscopy. At the late tetrad stage, WT anther wall was composed of five layers, including epidermis, endothecium, middle layer, parietal layer and tapetum from outside to inside, in which the tapetum cells exhibited highly condensed cytoplasm. Meanwhile, callose surrounding the tetrads in anther locule started to degenerate (Fig. 1e). Anthers at the same development stage in Arabidopsis and rice show similar cytological features except that their anther walls do not have a parietal layer (Sanders et al. 1999; Zhang et al. 2011). On the other hand, ms6 anthers at this stage had radically enlarged and highly vacuolated parietal and tapetal layers; in the locule, partially or non-separated irregular microspores with multiple nuclei were encased in callose, indicating that cytokinesis II of meiocytes was abnormal (Fig. 1f). By the time of the early pollen stage, enlarged pollens with thick wall were observed in the locule of WT anthers, and the anther wall was composed of an epidermis, an enlarged endothecium and a narrow parietal layer attached by tapetum remnants (Fig. 1g). Comparatively, in ms6 anthers, tapetum cells completely degraded and pollens collapsed while the parietal layer became abnormally vacuolated and swollen (Fig. 1h). These observations are similar to those of ms6 (Ames2), except that the tapetal layer of ms6 (Ames2) degenerated more rapidly. It almost completely degraded at the late tetrad stage (Ilarslan et al. 1999).
An SNP in Glyma.13G066600 is likely responsible for the male sterility in ms6
Previously, the ms6 mutation was mapped within a 3.72 Mb region on Chr13 between SSR markers Satt149 (Chr13:13,134,055 bp) and Satt030 (Chr13:16,855,019 bp) (Yang et al. 2014). To narrow down the region, WT and ms6 bulks, constructed from a BC5F2 mapping population originated from the cross of the ms6 mutant and ‘JiuB’, were subjected to the BSA-seq analysis. The ∆(SNP-index) plot showed that the ms6 mutation was associated with a 1.5 Mb region on Chr13 (Chr13:15,853,267–17,349,424 bp) (Fig. 2a), consisting to the previously reported interval (Yang et al. 2014). In addition, 214 SNP and 35 InDel variations were identified between two bulks in this region.
MS6 encodes an R2R3 MYB transcription factor a Manhattan plot of the SNP-index differences, ∆(SNP-index), between WT and ms6 bulks constructed with a BC5F2 mapping population derived from ms6 x ‘JiuB’. The red box highlights the region co-segregated with ms6. b Genetic linkage map of ms6. Genetic distances between two adjacent loci are indicated by centimorgans (cM). Chr 13, chromosome 13. c The MS6 gene structure and the mutated site in ms6. Black boxes represent the exons of MS6; arrow indicates the SNP location; nucleotide in red is the mutated site in ms6; underlined sequence is the MseI restriction site. d The GmTDF1-1 protein structure. Green blocks indicate the R2R3 MYB DNA-binding domain; arrow and amino acid in red indicate the amino acid substitution site in ms6. e Genotyping the plants with the CAPS marker based on the SNP between WT and ms6. The MseI-undigested (−) or digested ( +) PCR products are segregated on 4% agarose gel. WT, homozygous wild-type plant; Het, heterozygous (+ /ms6) plant; ms6, homozygous ms6 mutant. f Multiple sequence alignment of the R2 region from different MYB proteins. The residue corresponding to L76 in GmTDF1-1 (indicated by the red box) is conserved (color figure online)
Subsequently, a fine mapping was conducted with 328 individual plants in the BC5F2 mapping population with 9 polymorphic SSR markers identified in the 1.5 Mb interval, including BARCSOYSSR-13–0243, BARCSOYSSR-13–0244, BARCSOYSSR-13–0245, BARCSOYSSR-13–0249, BARCSOYSSR-13–0257, BARCSOYSSR-13–0259, BARCSOYSSR-13–0275, BARCSOYSSR-13–0277 and BARCSOYSSR-13–0283 (Fig. 2b; Fig. S1). Finally, the ms6 mutation was restricted to a 255 kb region (Chr13:16,428,596–16,683,664 bp) between BARCSOYSSR-13–0259 and BARCSOYSSR-13–0275 (Fig. 2b), which harbors 23 annotated genes. Among these genes, only Glyma.13G066600 showed a variation in the CDS region between WT and ms6 bulks according to the BSA-Seq results. It is a T-to-A SNP at position 330 in exon 2 of Glyma.13G066600, which destroys an MseI restriction site (Fig. 2c). Therefore, we verified this mutation by CAPS assay. A 126-bp region covering the SNP site was amplified by PCR from homozygous WT (+ / +), heterozygotes (+ /ms6) and homozygous ms6 (ms6/ms6), and the PCR products were subsequently digested with MseI. As expected, the WT amplicon was cleaved to a 99-bp band and a 27-bp band (invisible on the agarose gel); the ms6 amplicon only showed a 126-bp band because it could not be digested by MseI; the heterozygous amplicon had a 99-bp cut band and a 126-bp uncut band (Fig. 2e). Then, we further examined this position in 40 male-fertile wild, landrace and improved soybean accessions by the same CAPS assay, and they all exhibited the WT band pattern (Fig. S2, Table S2), indicating the position 330 of Glyma.13G066600 is highly conserved.
The SMART analysis indicated the protein encoded by Glyma.13G066600 is a transcription factor containing a typical R2R3 MYB DNA-binding domain at the N-terminal region (Fig. 2d). In the ms6 mutant, the SNP in Glyma.13G066600 leads to a L76H substitution (leucine at residue 76 substituted by histidine) in the polypeptide (Fig. 2d). Sequence comparison revealed L76 is well conserved in the R2 region of MYB proteins (Fig. 2f), implying it may be critical for the function of MYB proteins. The BLASTP analysis showed that Glyma.13G066600 encoded a putative homolog of TDF1, a key transcription factor regulating tapetum development and function (Zhu et al. 2008; Cai et al. 2015). The protein encoded by Glyma.13G066600 shares the highest sequence homology to AtTDF1 (44.9% identity and 64.6% similarity) in Arabidopsis and OsTDF1 (35.0% identity and 55.8% similarity) in rice (Fig. S3). Moreover, the null mutants of tdf1 in Arabidopsis and rice both promote vacuolization in tapetal cells and suppress the degradation of callose surrounding the tetrads, resulting in squeezed microspores and no-pollen phenotype (Zhu et al. 2008; Cai et al. 2015), which are similar to the phenotypes of ms6. These results together suggest that the T-to-A SNP in the CDS of Glyma.13G066600 is responsible for the male sterility of soybean ms6, and therefore we termed Glyma.13G066600 as MS6.
MS6 encodes a homolog of TDF1, an R2R3 MYB transcription factor only present in angiosperm
The above results indicated that the protein encoded by MS6 was likely a TDF1 homolog and named as GmTDF1-1 accordingly, although its sequence similarities shared with AtTDF1 and OsTDF1 were relatively low (Fig. S3). The relatively low homology was attributed to the highly diverged C-terminal region, which is a characteristic of MYB transcription factors (Jin and Martin 1999). Additionally, GmTDF1-1 exhibited a strong sequence homology (92% identity) to another soybean protein, which is encoded by Glyma.19G017900 and designated as GmTDF1-2 (Fig. S3). To verify the relationship between GmTDF1-1, GmTDF1-2 and TDF1, we conducted a phylogenetic analysis of GmTDF1-1, GmTDF1-2, AtTDF1, OsTDF1 and the putative homologs of TDF1 in other land plant species from different evolutionary lineages. AtMYB80 and OsMYB80 were used as outgroup sequences since MYB80 is the closest homologous protein of TDF1 (also known as MYB35) (Dubos et al. 2010).
As a result, these proteins are clustered into three groups (Fig. 3). In particular, AtTDF1, OsTDF1 and their putative homologs in other angiosperm species including GmTDF1-1 and GmTDF1-2 are clustered into a monophyletic group with two sub-branches. Specifically, AtTDF1 and all the TDF1 putative homologs in dicots are grouped in the same sub-branch with the one in the basal angiosperm A. trichopoda. OsTDF1 and all the TDF1 putative homologs in monocots including the basal or near basal monocots A. comosus and Z. marina are grouped in the other sub-branch (Fig. 3). Nevertheless, the TDF1 homologous proteins with the highest bit-score in lycophyte S. moellendorffii and moss P. patens form a second cluster with MYB80, and those in gymnosperm species, G. biloba and P. taeda, form the third cluster (Fig. 3). These data suggest that TDF1 is only present in angiosperm and has diverged at a very early stage in angiosperm evolution before monocots and dicots were differentiated. Moreover, the fact that GmTDF1-1 and GmTDF1-2 are placed together in the sub-branch of dicot TDF1s demonstrates that they are TDF1 homologs in soybean and their genes have arisen from a recent duplication event (Fig. 3).
Phylogenetic tree of GmTDF1-1 and TDF1 homologs from different plant species. The unrooted neighbor-joining phylogenetic tree based on GmTDF1-1 and TDF1 homologs is created by MEGA6. A. thaliana (Arabidopsis thaliana), A. trichopoda (Amborella trichopoda), A. comosus (Ananas comosus), G. biloba (Ginkgo biloba), G. max (Glycine max), M. acuminata (Musa acuminata), M. truncatula (Medicago truncatula), O. sativa (Oryza sativa), P. taeda (Pinus taeda), P. patens (Physcomitrella patens), S. bicolor (Sorghum bicolor), S. lycopersicum (Solanum lycopersicum), S. moellendorffii (Selaginella moellendorffii), V. vinifera (Vitis vinifera), Z. mays (Zea mays) and Z. marina (Zostera marina). Bootstrap values are indicated on the branches
L76H mutation abolished the ability of GmTDF1-1 to complement the male sterility of Arabidopsis attdf1 mutant
The function of TDF1 was likely conserved in angiosperm. Firstly, phylogenetic results showed that TDF1 homologs in angiosperm were clustered into a monophyletic group (Fig. 3). Secondly, depletion of TDF1 in Arabidopsis and rice caused similar detrimental effects on tapetum development (Zhu et al. 2008; Cai et al. 2015). Thirdly, expression of OsTDF1 driven by the native promoter AtTDF1p could rescue the male sterility of attdf1 (Cai et al. 2015). Therefore, to verify that GmTDF1-1 is a functional TDF1 critical for anther development and L76H mutation in ms6 disrupts the protein function, we conducted a complementary experiment by transferring the CDSs of AtTDF1, MS6, ms6 and GmTDF1-2 driven by AtTDF1p into attdf1.
As a result, most attdf1 plants transformed with AtTDF1 (14/20, rescued/transformants), MS6 (19/26, rescued/transformants) or GmTDF1-2 (8/10, rescued/transformants) were fully complemented, producing functional pollens and elongated siliques as wild type did (Fig. 4a–e, h–l, o–q). This demonstrated both GmTDF1-1 and GmTDF1-2 can substitute for AtTDF1 in Arabidopsis and they are functional TDF1s. By contrast, AtTDF1p:ms6 expressing GmTDF1-1L76H that carries L76H mutation (0/25, rescued/ transformants) failed to complement the attdf1’s sterility (Fig. 4a, b, f, h, i, m, r). Similarly, expression of AtTDF1p-driven AtTDF1L46H, which encodes a mutant form of AtTDF1 (AtTDF1L46H) corresponding to GmTDF1-1L76H, in attdf1 displayed the consistent result. All transformants (0/16, rescued/ transformants) developed just like attdf1 (Fig. 4a, b, g, h, i, n, s). On the other hand, overexpression (OE) of ms6 driven by 35S promoter in wild-type Arabidopsis did not affect anther development. We obtained 15 T1 transgenic OE lines, all producing normal pollens, in which three independent lines (ms6OE1 ~ 3) were shown in Fig. S4. These results strongly supported that the substitution of L76H abolishes the function of GmTDF1-1 and leads to the aberrant male development of ms6 plants. The results also demonstrated that leucine at position 76 in GmTDF1-1 (position 46 in AtTDF1) is crucial for the function of TDF1.
GmTDF1-1 rescues the fertility of attdf1 a–g Seed-setting staged col, attdf1 and transgenic plants in attdf1 background as labeled in each picture. Scale bar = 5 cm. h–n Alexander staining of the pollens from col, attdf1 and the corresponding transgenic plants placed above them. Scale bar = 100 μm. o–s Verification of the transgenic lines by RT-PCR and CAPS analyses. Top panels, RT-PCR analysis of the transgenes; Middle panels, RT-PCR analysis of AtActin as the native control; bottom panels, CAPS analysis showing the transgenic plants in attdf1 background
GmTDF1-1 is the major functional TDF1 in soybean
Complementary assay above showed that GmTDF1-1 and GmTDF1-2 both were functional TDF1 proteins, which raised the question of why ms6 exhibits male sterility when there is another TDF1 coding gene in the genome. To answer this question, we characterized the expression levels of MS6 and GmTDF1-2 genes by qRT-PCR in roots, stems, leaves, young flowers, pods and immature seeds. MS6 displayed a tissue-specific expression pattern with a much higher expression level in young flowers (Fig. 5). Then, the expression level of MS6 in different floral organs were further analyzed, which revealed MS6 is mainly expressed in anthers, similar to AtTDF1 and OsTDF1 (Fig. 5; Zhu et al. 2008; Cai et al. 2015). On the contrary, GmTDF1-2 is consistently expressed at a low level in all the examined tissues, indicating it is likely in the process of pseudogenization (Fig. 5). Different expression patterns of MS6 and GmTDF1-2 illustrated that GmTDF1-1 is the major TDF1 protein functioning in soybean anther development, and therefore the mutation at the MS6 locus leads to male sterility.
L76H mutation does not alter the subcellular localization or transactivation activity of GmTDF1-1
The subcellular localizations of GmTDF1-1 and GmTDF1-1L76H were analyzed by transiently expressing their GFP fusion proteins driven by 35S promoter in N. benthamiana leaves. Free GFP was also expressed as a control, which showed signals all over the cells (Fig. 6a). Comparatively, the fluorescence of GmTDF1-1-GFP and GmTDF1-1L76H-GFP was restricted in the nucleus, in agreement with the general subcellular distribution of transcription factors (Fig. 6a), and the result reveals that the L76H mutation did not affect the subcellular localization of GmTDF1-1.
GmTDF1-1 is located in the nucleus and possesses transactivation activity a Transient expression of GFP, GmTDF1-1-GFP and GmTDF1-1L76H -GFP in N. benthamiana leaves. For each protein, the images of GFP, chloroplast auto-fluorescence, bright field and merged signals are presented. Scale bar = 50 μm. b Transactivation analysis of GmTDF1-1, GmTDF1-1L76H and the DNA binding domain of GmTDF1-1 (GmTDF1-1DBD) in yeast. BD, the GAL4 DNA binding domain. BD alone was used as the negative control
TDF1 is a transcriptional activator in Arabidopsis and rice; therefore, GmTDF1-1 should possess transcription activation activity in soybean as well. We then performed a transactivation activity test in yeast (Fig. 6b). Yeast clones expressing GAL4 DNA binding domain (BD) only grew on the SD medium lack of tryptophan (SD/-Trp) but not on the selective medium (SD/-Trp-His) due to no transactivation activity in the BD region. A similar phenomenon was observed for the yeast clone expressing the BD-fused DNA binding domain of GmTDF1-1 (BD-GmTDF1-1DBD). In contrast, yeast clones expressing BD-GmTDF1-1 and BD-GmTDF1-1L76H grew well on both SD/-Trp and SD/-Trp-His mediums, showing the L76H substitution does not influence the transactivation activity of GmTDF1-1. As L76 is a conserved residue in the R2 region of the DNA binding domain (Fig. 2d and f) and L76H has no effect on protein’s subcellular location or transactivation activity, we suspected that the L76H mutation likely disrupted the function of GmTDF1-1 by altering its DNA binding capacity.
DYT1-TDF1-AMS-MYB80/MYB103/MS188-MS1 pathway downstream TDF1 is severely down regulated in the ms6 mutant
The above experiments showed that GmTDF1-1 is the major functional TDF1 homolog in soybean. In Arabidopsis, TDF1 is the critical component in a well-characterized genetic pathway regulating tapetal development and pollen wall formation. This pathway is composed of five transcription factors, which are two basic helix-loop-helix (bHLH) factors (DYSFUNCTIONAL TAPETUM 1 (DYT1) and ABORTED MICROSPORES (AMS)), two MYB factors (TDF1 and MYB80/MYB103/MS188) and one PHD-finger protein (MALE STERILITY 1 (MS1)) (Zhu et al. 2011; Lu et al. 2020). In this pathway, DYT1 directly activates the expression of TDF1, and TDF1 subsequently promotes the expression of AMS. Then, AMS is required for the expression of the gene encoding MYB80/MYB103/MS188, which is an activator critical for expressing MS1 (Fig. 7a). Depletion of any member in this cascade would lead to distorted tapetum and aborted pollens (Wilson et al. 2001; Sorensen et al. 2003; Zhang et al. 2006, 2007). The same regulatory cascade known as UDT1-TDF1-TDR-OsMS188-PTC1 was identified in rice as well (Cai et al. 2015).
The relative expression levels of anther development core regulators in WT and ms6 a Summarization of soybean genes encoding the TDF1-related regulatory pathway required for anther development. DYT1, TDF1, AMS1, MYB80/MYB103/MS188 and MS1 constitute the ordered transcription factor cascade identified in Arabidopsis. The genes encoding MS2 and A6 are well-known target genes of MYB80/MYB103/MS188 in Arabidopsis. Solid arrows indicate the direct activation; the dotted arrow indicates the indirect activation. b Relative expression levels of the above genes in WT and ms6 young flowers. The error bars denote ± SD calculated from three biological replicates, each with three-technique replicates. P values were calculated by using Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001)
Soybean ms6 exhibits similar cytological abnormalities to the null mutants, attdf1 and ostdf1, such as vacuolated tapetum cells, undissolved callose and crushed microspores (Fig. 1f, h; Ilarslan et al. 1999; Zhu et al. 2008; Cai et al. 2015). AtTDF1p-driven MS6 was able to recover the fertility of the attdf1 mutant as AtTDF1p-driven OsTDF1 did (Fig. 4d, k, p; Cai et al. 2015). These results suggested that TDF1’s function is conserved in Arabidopsis, rice and soybean, and implied that GmTDF1-1 controls anther development through the DYT1-TDF1-AMS-MYB80/MYB103/MS188-MS1 regulatory pathway as well. Through the homology search, the other four transcription factors in this pathway were also identified in soybean, in which DYT1 has four paralogs and the others have two paralogs (Fig. 7a). The expression levels of all the genes encoding the components of the pathway in WT and ms6 young flowers were assessed and compared by qRT-PCR analysis. Among the four genes encoding DYT1 homologs in soybean (GmDYT1s), the expression level of Glyma.13G250200 showed a significant increase in ms6 compared to that in WT, consisting to the previous report from Arabidopsis that TDF1 negatively feedback-regulates the expression of DYT1 who encodes the only transcription factor upstream TDF1 in the pathway (Cai et al. 2015). However, the expression levels of the other three genes encoding GmDYT1s were not significantly altered in ms6 (Fig. 7b), indicating the gene functions of GmDYT1s might have diverged. Moreover, the expressions levels of MS6 and GmTDF1-2 were not affected in ms6 (Fig. 7b), suggesting that soybean TDF1, GmTDF1-1, does not regulate the expressions of its own gene and its homolog GmTDF1-2.
On the other hand, expressions of the genes encoding the transcription factors downstream of TDF1 in the pathway, like AMS, MYB80/MYB103/MS188 and MS1, were mostly severely down regulated in ms6 compared to those in WT as expected (Fig. 7b). One exception was Glyma.01G047400 that encodes one of the MS1 homologs. It expressed similarly in WT and ms6 (Fig. 7b), implying that Glyma.01G047400 is functionally diverged from its paralog Glyma.02G107600 or even might lose the function in anther development. Moreover, we examined the expressions of two structural genes, A6 and MALE STERILE 2 (MS2), that are regulated by the transcription factor downstream of TDF1 (Zhang et al. 2007; Zhu et al. 2008; Wang et al. 2018). The A6 gene only has one homolog in soybean genome while MS2 has two (Fig. 7a), and the expressions of all three genes were strongly suppressed in ms6 flowers (Fig. 7b). A6 encodes a β-1,3-glucanase required to degrade callose (Hird et al. 1993), which explains why callose surrounding tetrads was not dissolved in ms6. MS2 encodes a fatty acyl reductase catalyzing the palmitoyl-acyl carrier protein into fatty alcohol, a precursor of the major component of pollen wall (Aarts et al. 1997; Chen et al. 2011b; Wang et al. 2018). In Arabidopsis, ms2 mutation disturbs the formation of pollen wall, which results in collapsed microspores and gives rise to no pollen anthers (Aarts et al. 1997). Similarly, no pollen is produced in anthers of soybean ms6 (Fig. 1d).
Taken together, in soybean ms6, the examined well-characterized downstream genes of TDF1 are severely down regulated, while expressions of the genes encoding TDF1 (GmTDF1-1 and GmTDF1-2) and the transcription factor upstream of TDF1 (DYT1) are not suppressed. It indicates that the DYT1-TDF1-AMS-MYB80/MYB103/MS188-MS1 genetic pathway is likely present in soybean and functions conservatively to assure anther development. It also reflects that the DYT1-TDF1-AMS-MYB80/MYB103/MS188-MS1 genetic pathway is blocked at the step regulated by TDF1 in the ms6 mutant, demonstrating that the mutation of Glyma.13G066600/MS6 that encodes the major TDF1 in soybean is the determinant for the male sterility of ms6.
Discussion
Plant male sterile mutants are important materials for studying anther development and crucial tools for crop hybrid breeding. So far, 13 genetic loci have been reported to condition the male sterile phenotype in soybean, including ms1-ms9, msp, msMOS, mst-M and msNJ (Yang et al. 2014; Zhao et al. 2019; Nie et al. 2019; Thu et al. 2019). Nevertheless, only ms4 has been molecularly identified, which is caused by the mutation of Glyma.02G243200, a gene encoding a PHD-finger protein involved in the meiosis process of microsporocyte (Thu et al. 2019). In the present study, we cloned another male sterile gene, MS6, by characterizing the ms6 allele maintained in the germplasm T295H. Results from genetic mapping, comparative high-throughput sequencing, conservation study, phylogenetic analysis, complementary experiment, gene expression patterns and differential expressions of genes in the TDF1-involved regulatory pathway all lead to the conclusion that the male sterility of ms6 from T295H is caused by the missense mutation at the Glyma.13G066600 locus (MS6), which encodes a soybean homolog of TDF1, GmTDF1-1 (Figs. 2-5, 7). TDF1 is a well-known R2R3 MYB transcription factor required for appropriate tapetum development (Zhu et al. 2008; Cai et al. 2015). The missense mutation in the ms6 allele results in the substitution of a highly conserved leucine residue by a histidine residue at the position 76 (L76H) in the R2 region of GmTDF1-1 (Fig. 2). The mutation abolishes the function of GmTDF1-1 (Fig. 4), likely by interfering its DNA-binding activity, because the transactivation activity and subcellular distribution of GmTDF1-1L76H encoded by the ms6 allele are not disturbed (Fig. 6) and the ms6 allele is expressed at a similar level to that in WT (Fig. 7). Phylogenetic and complementation analyses showed that GmTDF1-1 has a recently diverged and functional paralog, GmTDF1-2 (Figs. 3, 4). However, the GmTDF1-2 gene is expressed constitutively at a very low level; therefore, it may only have a trivial contribution to the anther development if there is any, and its presence cannot compensate for the deficiency of MS6 (Fig. 5).
TDF1 is conservatively present in angiosperm species (Fig. 3), regulating the tapetal and microspore development (Zhu et al. 2008; Cai et al. 2015). The major TDF1-involved genetic pathway is the ordered transcription factor cascade identified as DYT1-TDF1-AMS-MYB80/MYB103/MS188-MS1 in Arabidopsis (Zhu et al. 2011; Lu et al. 2020) and UDT1-TDF1-TDR-OsMS188-PTC1 in rice (Cai et al. 2015). Expressions of rice and soybean TDF1s (OsTDF1, GmTDF1-1 and GmTDF1-2) in Arabidopsis attdf1 mutant under the control of the native AtTDF1 promoter can recover the fertility of the mutant plants, indicating that TDF1’s major functions are quite conserved (Fig. 4; Cai et al. 2015). However, slight divergences of TDF1 genes are present in different species. For example, in situ hybridization showed that AtTDF1 was expressed strongly and equivalently in tapetum and meiocytes at stage 6, while OsTDF1 was expressed much stronger in tapetum than in meiocytes at the similar development stage (Zhu et al. 2008, 2011; Cai et al. 2015). Expressing OsTDF1 in attdf1 only partially recovered the expression levels of downstream target genes like AMS, MYB80/MYB103/MS188 and MS1 (Zhu et al. 2008, 2011; Cai et al. 2015). The function of the major TDF1 in soybean (MS6) is likely more diverged. For instance, mutants attdf1 and ostdf1 process meiosis successfully to generate well-separated tetrads, while both soybean ms6 mutants (Ames1 and Ames2) produce partially- or non-separated multi-nucleic tetrads (Fig. 1f; Skorupska and Palmer 1989; Ilarslan et al. 1999). Additionally, compared to Arabidopsis and rice, soybean possesses an extra anther wall layer between the tapetal and middle layers, termed as the parietal layer (Fig. 1; Ilarslan et al. 1999). In ms6, the parietal layer is vacuolated and obsessively enlarged, indicating that MS6 also plays an important role in regulating the parietal layer’s development. To dissect the function of GmTDF1-1 in soybean anther development, future investigation of the GmTDF1-1 downstream network by using RNA-Seq and ChIP-seq technologies is needed.
Stable GMS mutants can facilitate crop improvement via the canonical recurrent selection strategy (Lewers et al. 1996) or the novel GMS-based hybrid breeding system, SPT (Perez-Prat and van Lookeren Campagne 2002; Weber et al. 2009). The main idea of the SPT technology is to create a hemizygous transgenic maintainer line by introducing a resistance gene (for screening transgenic lines) and a special gene cluster (for producing viable ms pollens) to a recessive sporophytic ms mutant. The gene cluster is composed of at least three fundamental genes (Wu et al. 2016). The first one is the wild-type MS gene for rescuing ms’s detrimental effects on anther sporophytic cells. The 2nd one is a male gametophyte-killer gene for killing the male gametes carrying the transgenic component, and therefore only non-transgenic ms pollens are viable for fertilization and seed production (Chang et al. 2016; Song et al. 2021). The 3rd one is a phenotypic reporter gene for monitoring the purity of the propagated ms seeds, such as fluorescence gene expressed in the aleurone layer of seeds (Chang et al. 2016; Zhang et al. 2018b) or anthocyanin synthesis gene expressed in young seedlings (Du et al. 2020). The SPT system broadens the germplasm choices of parental lines to breed hybrids with superior heterosis, reduces the risk caused by weather changes and is regarded as the third generation of hybrid technology (Song et al. 2021).
Therefore, the recessive sporophytic ms mutants conditioned by well-characterized MS genes are critical for developing the SPT system. However, compared to the major crops like rice and maize with dozens of ms mutants controlled by different loci (Guo and Liu 2012; Wan et al. 2019), soybean only has 13 ms loci reported (Yang et al. 2014; Zhao et al. 2019; Nie et al. 2019; Thu et al. 2019). One major reason is that soybean is a paleopolyploid with two recent rounds of whole-genome duplication (WGD) occurring ~ 13 and ~ 59 million years ago, and about 75% of the genes exist with multiple copies (Schmutz et al. 2010). For example, among the genes we investigated in the present study, all but A6 have ≥ 2 paralogs in soybean genome (Fig. 7a). Therefore, there is a big chance that spontaneous mutation at one anther-development-related gene would not affect male gametogenesis due to gene redundancy. In hence, the ms mutants are hardly achieved in soybean through traditional random-mutagenesis approaches. Among the reported ms mutants, sporophytic ms6 is an ideal genetic material for the SPT technology due to its stable no pollen phenotype (Fig. 1c, d; Skorupska and Palmer 1989; Ilarslan et al. 1999). Identification of the MS6 gene in the present study provides an essential element in establishing the potential ms6-based SPT system for soybean hybrid-seed production. Elucidation of the mutation site at the ms6 allele also provides an accurate marker to facilitate soybean improvement via recurrent selection.
References
Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12:615–623. https://doi.org/10.1046/j.1365-313x.1997.00615.x
Bassam BJ, Caetano-Anollés G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83. https://doi.org/10.1016/0003-2697(91)90120-i
Cai CF, Zhu J, Lou Y, Guo ZL, Xiong SX, Wang K, Yang ZN (2015) The functional analysis of OsTDF1 reveals a conserved genetic pathway for tapetal development between rice and Arabidopsis. Sci Bull 60:1073–1082. https://doi.org/10.1007/s11434-015-0810-3
Chang Z, Chen Z, Wang N, Xie G, Lu J, Yan W, Zhou J, Tang X, Deng XW (2016) Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc Natl Acad Sci U S A 113:14145–14150. https://doi.org/10.1073/pnas.1613792113
Chen DC, Yang BC, Kuo TT (1992) One-step transformation of yeast in stationary phase. Curr Genet 21:83–84. https://doi.org/10.1007/BF00318659
Chen L, Lei D, Tang W, Xiao Y (2011a) Thoughts and practice on some problems about research and application of two-line hybrid rice. Rice Sci 18:79–85. https://doi.org/10.1016/S1672-6308(11)60012-7
Chen W, Yu XH, Zhang K, Shi J, Oliveira SD, Schreiber L, Shanklin J, Zhang D (2011b) Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol 157:842–853. https://doi.org/10.1104/pp.111.181693
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581. https://doi.org/10.1016/j.tplants.2010.06.005
Du M, Zhou K, Liu Y, Deng L, Zhang X, Lin L, Zhou M, Zhao W, Wen C, Xing J, Li CB, Li C (2020) A biotechnology-based male-sterility system for hybrid seed production in tomato. Plant J 102:1090–1100. https://doi.org/10.1111/tpj.14678
Guo JX, Liu YG (2012) Molecular control of male reproductive development and pollen fertility in rice. J Integr Plant Biol 54:967–978. https://doi.org/10.1111/j.1744-7909.2012.01172.x
Hird DL, Worrall D, Hodge R, Smartt S, Paul W, Scott R (1993) The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to β-1,3-glucanases. Plant J 4:1023–1033. https://doi.org/10.1046/j.1365-313x.1993.04061023.x
Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. https://doi.org/10.1016/0378-1119(89)90358-2
Ilarslan H, Horner HT, Palmer RG (1999) Genetics and cytology of a new male-sterile, female-fertile soybean mutant. Crop Sci 39:58–64. https://doi.org/10.2135/cropsci1999.0011183X003900010009x
Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585. https://doi.org/10.1023/a:1006319732410
Kim YJ, Zhang D (2018) Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci 23:53–65. https://doi.org/10.1016/j.tplants.2017.10.001
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181. https://doi.org/10.1016/0888-7543(87)90010-3
Levings CS 3rd (1990) The Texas cytoplasm of maize: cytoplasmic male sterility and disease susceptibility. Science 250:942–947. https://doi.org/10.1126/science.250.4983.942
Lewers KS, Martin SKS, Widrlechner MP, Palmer RG, Hedges BR (1996) Hybrid soybean seed production: comparison of three methods. Crop Sci 36:1560–1567. https://doi.org/10.2135/cropsci1996.0011183X003600060026x
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆Ct method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lu JY, Xiong SX, Yin W, Teng XD, Lou Y, Zhu J, Zhang C, Gu JN, Wilson ZA, Yang ZN (2020) MS1, a direct target of MS188, regulates the expression of key sporophytic pollen coat protein genes in arabidopsis. J Exp Bot 71:4877–4889. https://doi.org/10.1093/jxb/eraa219
Nie Z, Zhao T, Liu M, Dai J, He T, Lyu D, Zhao J, Yang S, Gai J (2019) Molecular mapping of a novel male-sterile gene msNJ in soybean [Glycine max (L.) Merr.]. Plant Reprod 32:371–380. https://doi.org/10.1007/s00497-019-00377-6
Perez-Prat E, van Lookeren Campagne MM (2002) Hybrid seed production and the challenge of propagating male-sterile plants. Trends Plant Sci 7:199–203. https://doi.org/10.1016/s1360-1385(02)02252-5
Peterson R, Slovin JP, Chen C (2010) A simplified method for differential staining of aborted and non-aborted pollen grains. Int J Plant Biol 1:e13. https://doi.org/10.4081/pb.2010.e13
Palmer RG, Gai J, Sun H, Burton JW (2001) Production and evaluation of hybrid soybean. In: J. Janick (ed) Plant breeding reviews, Vol 21. John Wiley & Sons, New York, pp 263-307. https://doi.org/10.1002/9780470650196.ch7
Palmer RG, Holland JB, Lewers KS (1998) Recombination values for the Ms6-W1 chromosome region in different genetic backgrounds in soybean. Crop Sci 38:293–296. https://doi.org/10.2135/cropsci1998.0011183X003800020002x
Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11:297–322. https://doi.org/10.1007/s004970050158
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the paleopolyploid soybean. Nature 463:178–183. https://doi.org/10.1038/nature08670
Skorupska H, Palmer RG (1989) Genetics and cytology of the ms6 male-sterile soybean. J Hered 80:304–310. https://doi.org/10.1093/oxfordjournals.jhered.a110858
Song S, Wang T, Li Y, Hu J, Kan R, Qiu M, Deng Y, Liu P, Zhang L, Dong H, Li C, Yu D, Li X, Yuan D, Yuan L, Li L (2021) A novel strategy for creating a new system of third-generation hybrid rice technology using a cytoplasmic sterility gene and a genic male-sterile gene. Plant Biotechnol J 19:251–260. https://doi.org/10.1111/pbi.13457
Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33:413–423. https://doi.org/10.1046/j.1365-313x.2003.01644.x
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183. https://doi.org/10.1111/tpj.12105
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Thu SW, Rai KM, Sandhu D, Rajangam A, Balasubramanian VK, Palmer RG, Mendu V (2019) Mutation in a PHD-finger protein MS4 causes male sterility in soybean. BMC Plant Biol 19:378. https://doi.org/10.1186/s12870-019-1979-4
Wang K, Guo ZL, Zhou WT, Zhang C, Zhang ZY, Lou Y, Xiong SX, Yao XZ, Fan JJ, Zhu J, Yang ZN (2018) The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation. Plant Physiol 178:283–294. https://doi.org/10.1104/pp.18.00219
Wan X, Wu S, Li Z, Dong Z, An X, Ma B, Tian Y, Li J (2019) Maize genic male-sterility genes and their applications in hybrid breeding: progress and perspectives. Mol Plant 12:321–342. https://doi.org/10.1016/j.molp.2019.01.014
Weber N, Commuri P, Rood T, Townsend R (2009) Petition for the determination of nonregulated status for Maize 32138 SPT maintainer used in the Pioneer Seed Production Technology (SPT) process. Submitted to the USDA-APHIS by Pioneer Hi-Bred International, Inc. Available at: http:// www.aphis.usda.gov/brs/aphisdocs/08_33801p.pdf (last accessed 13 August 2015)
Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ (2001) The Arabidopsis MALE STERILITY 1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28:27–39. https://doi.org/10.1046/j.1365-313x.2001.01125.x
Wu Y, Fox TW, Trimnell MR, Wang L, Xu RJ, Cigan AM, Huffman GA, Garnaat CW, Hershey H, Albertsen MC (2016) Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol J 14:1046–1054. https://doi.org/10.1111/pbi.12477
Yang Y, Speth BD, Boonyoo N, Baumert E, Atkinson TR, Palmer RG, Sandhu D (2014) Molecular mapping of three male-sterile, female-fertile mutants and generation of a comprehensive map of all known male sterility genes in soybean. Genome 57:155–160. https://doi.org/10.1139/gen-2014-0018
Zhang D, Luo X, Zhu L (2011) Cytological analysis and genetic control of rice anther development. J Genet Genomics 38:379–390. https://doi.org/10.1016/j.jgg.2011.08.001
Zhang D, Chang E, Yu X, Chen Y, Yang Q, Cao Y, Li X, Wang Y, Fu A, Xu M (2018a) Molecular characterization of magnesium chelatase in Soybean [Glycine max (L.) Merr.]. Front Plant Sci 9:720. https://doi.org/10.3389/fpls.2018.00720
Zhang D, Wu S, An X, Xie K, Dong Z, Zhou Y, Xu L, Fang W, Liu S, Liu S, Zhu T, Li J, Rao L, Zhao J, Wan X (2018b) Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol J 16:459–471. https://doi.org/10.1111/pbi.12786
Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133:3085–3095. https://doi.org/10.1242/dev.02463
Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, Huang H, Xia HJ, Yang ZN (2007) Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J 52:528–538. https://doi.org/10.1111/j.1365-313X.2007.03254.x
Zhao L, Sun H, Wang S, Wang Y, Huang M, Li J (2004) Breeding of hybrid soybean HybSoy1. Chinese J Oil Crop Sci 26:15–17 (in Chinese)
Zhao Q, Tong Y, Yang C, Yang Y, Zhang M (2019) Identification and mapping of a new soybean male-sterile gene, mst-M. Front Plant Sci 10:94. https://doi.org/10.3389/fpls.2019.00094
Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang Z (2008) Defective in Tapetal Development and Function 1 is essential for anther development and tapetal function for microspore maturation in arabidopsis. Plant J 55:266–277. https://doi.org/10.1111/j.1365-313X.2008.03500.x
Zhu J, Lou Y, Xu X, Yang ZN (2011) A genetic pathway for tapetum development and function in arabidopsis. J Integr Plant Biol 53:892–900. https://doi.org/10.1111/j.1744-7909.2011.01078.x
Acknowledgements
We thank U.S. National Plant Germplasm System (NPGS) for providing T295H and other U.S. germplasms. We thank Dr.s Zhiping Huang and Dagang Wang from Anhui Academy of Agricultural Sciences, Anhui, China for providing cultivated soybeans originated in Anhui province. We thank Dr. Mengjuan Liu from Northwest A&F University, Yangling, China for providing other Chinese soybean cultivars. We thank Dr.s Yingshan Dong and Xiaodong Liu from Jilin Academy of Agricultural Sciences, Jilin, China for providing wild soybean germplasms. Finally, we thank Dr. Zhongnan Yang from Shanghai Normal University, Shanghai, China for supporting the attdf1 heterozygotes.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0101500 and 2016YFD0101503 to MX), the National Natural Science Foundation of China (32000598 to JY; 31971969 to CZ), Jilin Provincial Agricultural Science and Technology Innovation Project (CXGC2021ZD002 to CZ) and Shaanxi Provincial Natural Science Foundation (2020JQ-578 to JY).
Author information
Authors and Affiliations
Contributions
JY, MX and CZ designed this project. LZ and CZ developed the BC5F2 segregation population. GZ, YZ and CZ engaged in the BSA-Seq and genetic mapping work. JY, WL, PW, AF and MX were involved in the rest of the experiments in this study. JY, GZ, WL, AF, CZ and MX wrote the paper. All the authors revised the paper.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declared with no conflicts and approved the manuscript.
Additional information
Communicated by Istvan Rajcan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, J., Zhao, G., Li, W. et al. A single nucleotide polymorphism in an R2R3 MYB transcription factor gene triggers the male sterility in soybean ms6 (Ames1). Theor Appl Genet 134, 3661–3674 (2021). https://doi.org/10.1007/s00122-021-03920-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03920-0