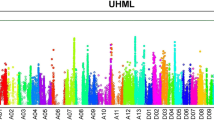
Abstract
Key message
Genetic variation in a G. barbadense population was revealed using resquencing. GWAS on G.barbadense population identified several candidate genes associated with fiber strength and lint percentage.
Abstract
Gossypium barbadense is the second-largest cultivated cotton species planted in the world, which is characterized by high fiber quality. Here, we described the global pattern of genetic polymorphisms for 240 G. barbadense accessions based on the whole-genome resequencing. A total of 3,632,231 qualified single-nucleotide polymorphisms (SNPs) and 221,354 insertion–deletions (indels) were obtained. We conducted a genome-wide association study (GWAS) on 12 traits under four environments. Two traits with more stable associated variants, fiber strength and lint percentage, were chosen for further analysis. Three putative candidate genes, HD16 orthology (GB_D11G3437), WDL2 orthology (GB_D11G3460) and TUBA1 orthology (GB_D11G3471), on chromosome D11 were found to be associated with fiber strength, and one gene orthologous to Arabidopsis Receptor-like protein kinase HERK 1 (GB_A07G1034) was predicated to be the candidate gene for the lint percentage improvement. The identified genes may serve as promising targets for genetic engineering to accelerate the breeding process for G. barbadense and the high-density genome variation map constructed in this work may facilitate our understanding of the genetic architecture of cotton traits.




Similar content being viewed by others
Data availability
The genomic resequencing datasets generated during the current study are available in the NCBI Sequence Read Archive repository under accession number PRJNA728217.
References
Abdelraheem A, Fang DD, Dever J, Zhang JF (2020) QTL analysis of agronomic, fiber quality, and abiotic stress tolerance traits in a recombinant inbred population of pima cotton. Crop Sci 60:1823–1843. https://doi.org/10.1002/csc2.20153
Abe T, Hashimoto T (2005) Altered microtubule dynamics by expression of modified alpha-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. Plant J 43:191–204. https://doi.org/10.1111/j.1365-313X.2005.02442.x
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664. https://doi.org/10.1101/gr.094052.109
Alifu A, Zhu J, Li J, Wang W, Ning X, Liu Z et al (2020) Progress, problems and prospects of xinjiang long-staple cotton (G. barbadense L.) breeding. Xinjiang Agric Sci 57:393–400
Chee P, Draye X, Jiang CX, Decanini L, Delmonte TA, Bredhauer R et al (2005) Molecular dissection of interspecific variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: I. Fiber Elongation Theor Appl Genet 111:757–763. https://doi.org/10.1007/s00122-005-2063-z
Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang TZ, Guo WZ et al (2007) Toward sequencing cotton (Gossypium) genomes. Plant Physiol 145:1303–1310. https://doi.org/10.1104/pp.107.107672
Chen Y, Liu GD, Ma HH, Song ZQ, Zhang CY, Zhang JX et al (2018) Identification of introgressed alleles conferring high fiber quality derived from Gossypium barbadense L. in secondary mapping populations of G. hirsutum L. Front. Plant Sci. https://doi.org/10.3389/fpls.2018.01023
Chen ZJ, Sreedasyam A, Ando A, Song QX, De Santiago LM, Hulse-Kemp AM et al (2020) Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat Genet 52:525–533. https://doi.org/10.1038/s41588-020-0614-5
Dai C, Xue HW (2010) Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J 29:1916–1927. https://doi.org/10.1038/emboj.2010.75
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al (2011) The variant call format and VCFtools. Bioinform 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Du X, Huang G, He S, Yang Z, Sun G, Ma X et al (2018) Resequencing of 243 diploid cotton accessions based on an updated a genome identifies the genetic basis of key agronomic traits. Nat Genet 50:796–802. https://doi.org/10.1038/s41588-018-0116-x
Fan LP, Wang LP, Wang XY, Zhang HY, Zhu YF, Guo JY et al (2018) A high-density genetic map of extra-long staple cotton (Gossypium barbadense) constructed using genotyping-by-sequencing based single nucleotide polymorphic markers and identification of fiber traits-related QTL in a recombinant inbred line population. BMC Genomics 19:12. https://doi.org/10.1186/s12864-018-4890-8
Fang L, Wang Q, Hu Y, Jia Y, Chen J, Liu B et al (2017) Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat Genet 49:1089–1098. https://doi.org/10.1038/ng.3887
Fang L, Zhao T, Hu Y, Si ZF, Zhu XF, Han ZG et al (2021) Divergent improvement of two cultivated allotetraploid cotton species. Plant Biotechnol J. https://doi.org/10.1111/pbi.13547
Gross SD, Anderson RA (1998) Casein kinase I: Spatial organization and positioning of a multifunctional protein kinase family. Cell Signal 10:699–711. https://doi.org/10.1016/s0898-6568(98)00042-4
Hao J, Tu LL, Hu HY, Tan JF, Deng FL, Tang WX et al (2012) GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J Exp Bot 63:6267–6281. https://doi.org/10.1093/jxb/ers278
Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y et al (2019) Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet 51:739–748. https://doi.org/10.1038/s41588-019-0371-5
Huang DB, Wang SG, Zhang BC, Shang-Guan KK, Shi YY, Zhang DM et al (2015) A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27:1681–1696. https://doi.org/10.1105/tpc.15.00015
Iqbal N, Manalil S, Chauhan BS, Adkins SW (2019) Glyphosate-tolerant cotton in Australia: successes and failures. Arch Agron Soil Sci 65:1536–1553. https://doi.org/10.1080/03650340.2019.1566720
Lacape J-M, Llewellyn D, Jacobs J, Arioli T, Becker D, Calhoun S et al (2010) Meta-analysis of cotton fiber quality QTLs across diverse environments in a Gossypium hirsutum x G. barbadense RIL population. BMC Plant Biol. https://doi.org/10.1186/1471-2229-10-1
Li H, Durbin R (2010) Fast and accurate long-read alignment with burrows-wheeler transform. Bioinform 26:589–595. https://doi.org/10.1093/bioinformatics/btp698
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al (2009) The sequence alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Liu HJ, Yan JB (2019) Crop genome-wide association study: a harvest of biological relevance. Plant J 97:8–18. https://doi.org/10.1111/tpj.14139
Ma Z, He S, Wang X, Sun J, Zhang Y, Zhang G et al (2018) Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat Genet 50:803–813. https://doi.org/10.1038/s41588-018-0119-7
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al (2010) The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Nie XH, Tu JL, Wang B, Zhou XF, Lin ZX (2015) A BIL population derived from G hirsutum and G barbadense provides a resource for cotton genetics and breeding. PLoS ONE 10:14. https://doi.org/10.1371/journal.pone.0141064
Nie XH, Wen TW, Shao PX, Tang BH, Nuriman-guli A, Yu Y et al (2020) High-density genetic variation maps reveal the correlation between asymmetric interspecific introgressions and improvement of agronomic traits in upland and pima cotton varieties developed in Xinjiang, China. Plant J 103:677–689. https://doi.org/10.1111/tpj.14760
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin DC et al (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–417. https://doi.org/10.1038/nature11798
Percy RG, Turcotte EL (1991) Early-maturing, short-statured American Pima cotton parents improve agronomic traits of interspecific hybrids. Crop Sci 31:709–712. https://doi.org/10.2135/cropsci1991.0011183X003100030033x
Percy RG, Turcotte EL (1992) Interspecific hybrid fiber characteristics of cotton altered by unconventional Gossypium barbadense L. fiber genotypes. Crop Sci 32:1437–1441. https://doi.org/10.2135/cropsci1992.0011183X003200060025x
Percy RG, Wendel JF (1990) Allozyme evidence for the origin and diversification of Gossypium barbadense L. Theor Appl Genet 79:529–542. https://doi.org/10.1007/bf00226164
Perrin RM, Wang Y, Yuen CYL, Will J, Masson PH (2007) WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. Plant J 49:961–971. https://doi.org/10.1111/j.1365-313X.2006.03015.x
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:6. https://doi.org/10.1093/nar/29.9.e45
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
Shi Y, Li W, Li A, Ge R, Zhang B, Li J et al (2015) Constructing a high-density linkage map for Gossypium hirsutum x Gossypium barbadense and identifying QTLs for lint percentage. J Integr Plant Biol 57:450–467. https://doi.org/10.1111/jipb.12288
Su XJ, Zhu GZ, Song XH, Xu HJ, Li WX, Ning XZ et al (2020) Genome-wide association analysis reveals loci and candidate genes involved in fiber quality traits in sea island cotton (Gossypium barbadense). BMC Plant Biol 20:11. https://doi.org/10.1186/s12870-020-02502-4
Varshney RK, Pandey MK, Bohra A, Singh VK, Thudi M, Saxena RK (2019) Toward the sequence-based breeding in legumes in the post-genome sequencing era. Theor Appl Genet 132:797–816. https://doi.org/10.1007/s00122-018-3252-x
Wang K, Li MY, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:7. https://doi.org/10.1093/nar/gkq603
Wang P, Zhu YJ, Song LL, Cao ZB, Ding YZ, Liu BL et al (2012) Inheritance of long staple fiber quality traits of Gossypium barbadense in G. hirsutum background using CSILs. Theor Appl Genet 124:1415–1428. https://doi.org/10.1007/s00122-012-1797-7
Wang MJ, Tu LL, Lin M, Lin ZX, Wang PC, Yang QY et al (2017) Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat Genet 49:579–587. https://doi.org/10.1038/ng.3807
Wang F, Zhang J, Chen Y, Zhang C, Gong J, Song Z et al (2020a) Identification of candidate genes for key fibre-related QTLs and derivation of favourable alleles in Gossypium hirsutum recombinant inbred lines with G. barbadense introgressions. Plant Biotechnol J 18:707–720. https://doi.org/10.1111/pbi.13237
Wang L, Wang G, Long L, Altunok S, Feng Z, Wang D et al (2020b) Understanding the role of phytohormones in cotton fiber development through omic approaches; recent advances and future directions. Int J Biol Macromol 163:1301–1313. https://doi.org/10.1016/j.ijbiomac.2020.07.104
Wen TW, Dai BS, Wang T, Liu XX, You CY, Lin ZX (2019) Genetic variations in plant architecture traits in cotton (Gossypium hirsutum) revealed by a genome-wide association study. Crop J 7:209–216. https://doi.org/10.1016/j.cj.2018.12.004
Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78:139–186. https://doi.org/10.1016/S0065-2113(02)78004-8
Westengen O, Huaman Z, Heun M (2005) Genetic diversity and geographic pattern in early South American cotton domestication. Theor Appl Genet 110:392–402. https://doi.org/10.1007/s00122-004-1850-2
Yang JA, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82. https://doi.org/10.1016/j.ajhg.2010.11.011
Zhang Z, Ruan Y-L, Zhou N, Wang F, Guan X, Fang L et al (2017) Suppressing a putative sterol carrier gene reduces plasmodesmal permeability and activates sucrose transporter genes during cotton fiber elongation. Plant Cell 29:2027–2046. https://doi.org/10.1105/tpc.17.00358
Zhang Z, Li J, Jamshed M, Shi Y, Liu A, Gong J et al (2020) Genome-wide quantitative trait loci reveal the genetic basis of cotton fibre quality and yield-related traits in a Gossypium hirsutum recombinant inbred line population. Plant Biotechnol J 18:239–253. https://doi.org/10.1111/pbi.13191
Zhou X, Stephens M (2012) Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44:821-U136. https://doi.org/10.1038/ng.2310
Funding
We are grateful to Xinjiang Tarim River Seed Co., Ltd. for kindly helping us with the investigation works of traits in the field. This work was supported by the National Key Technology R&D program of China (2016YFD0101404), China Agriculture Research System (CARS-18–25) and Jiangsu Collaborative Innovation Center for Modern Crop Production.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare they have no conflicts of interest.
Additional information
Communicated by David D Fang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, J., Hui, Y., Chen, J. et al. Whole-genome resequencing of 240 Gossypium barbadense accessions reveals genetic variation and genes associated with fiber strength and lint percentage. Theor Appl Genet 134, 3249–3261 (2021). https://doi.org/10.1007/s00122-021-03889-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03889-w