Abstract
Key message
Polygenic genome-wide association mapping identified two regions of the cowpea genome associated with different components of resistance to its major post-harvest pest, the seed beetle Callosobruchus maculatus.
Abstract
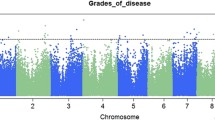
Cowpea (Vigna unguiculata) is an important grain and fodder crop in arid and semi-arid regions of Africa, Asia, and South America, where the cowpea seed beetle, Callosobruchus maculatus, is a serious post-harvest pest. Development of cultivars resistant to C. maculatus population growth in storage could increase grain yield and quality and reduce reliance on insecticides. Here, we use a MAGIC (multi-parent, advanced-generation intercross) population of cowpea consisting of 305 recombinant inbred lines (RILs) to identify genetic variants associated with resistance to seed beetles. Because inferences regarding the genetic basis of resistance may depend on the source of the pest or the assay protocol, we used two divergent geographic populations of C. maculatus and two complementary assays to measure several aspects of resistance. Using polygenic genome-wide association mapping models, we found that the cowpea RILs harbor substantial additive-genetic variation for most resistance measures. Variation in several components of resistance, including larval development time and survival, was largely explained by one or several linked loci on chromosome 5. A second region on chromosome 8 explained increased seed resistance via the induction of early-exiting larvae. Neither of these regions contained genes previously associated with resistance to insects that infest grain legumes. We found some evidence of gene–gene interactions affecting resistance, but epistasis did not contribute substantially to resistance variation in this mapping population. The combination of mostly high heritabilities and a relatively consistent and simple genetic architecture increases the feasibility of breeding for enhanced resistance to C. maculatus.







Similar content being viewed by others
References
Amusa OD, Ogunkanmi LA, Adetumbi JA, Akinyosoye ST, Ogundipe OT (2018) Genetics of bruchid (Callosobruchus maculatus Fab.) resistance in cowpea (Vigna unguiculata (L.) Walp.). J Stored Prod Res 75:18–20
Appleby J, Credland P (2003) Variation in responses to susceptible and resistant cowpeas among west African populations of Callosobruchus maculatus (Coleoptera: Bruchidae). J Econ Entomol 96(2):489–502
Asif M, Rooney LW, Ali R, Riaz MN (2013) Application and opportunities of pulses in food system: a review. Crit Rev Food Sci Nutr 53(11):1168–1179
Boeke SJ, Van Loon JJ, Van Huis A, Dicke M (2004) Host preference of Callosobruchus maculatus: a comparison of life history characteristics for three strains of beetles on two varieties of cowpea. J Appl Entomol 128(6):390–396
Boukar O, Belko N, Chamarthi S, Togola A, Batieno J, Owusu E, Haruna M, Diallo S, Umar ML, Olufajo O et al (2019) Cowpea (Vigna unguiculata): Genetics, genomics and breeding. Plant Breed 138(4):415–424
Boukar O, Abberton M, Oyatomi O, Togola A, Tripathi L, Fatokun C (2020) Introgression breeding in cowpea [Vigna unguiculata (L.) Walp.]. Frontiers in Plant Science 11:567425
Cavanagh C, Morell M, Mackay I, Powell W (2008) From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr Opinion Plant Biol 11(2):215–221
Chotechung S, Somta P, Chen J, Yimram T, Chen X, Srinives P (2016) A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: Bruchidae) resistance in mungbean (Vigna radiata). Theor Appl Genet 129(9):1673–1683
Crawford L, Zeng P, Mukherjee S, Zhou X (2017) Detecting epistasis with the marginal epistasis test in genetic mapping studies of quantitative traits. PLoS Genet 13(7):e1006869
Cruz LP, de Sá LF, Santos LA, Gravina GA, Carvalho AO, Fernandes KVS, Freire Filho FR, Gomes VM, Oliveira AEA (2016) Evaluation of resistance in different cowpea cultivars to Callosobruchus maculatus infestation. J Pest Sci 89(1):117–128
De Lorenzo G, D’Ovidio R, Cervone F (2001) The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu Rev Phytopathol 39(1):313–335
Deshpande V, Makanur B, Deshpande S, Adiger S, Salimath P (2011) Quantitative and qualitative losses caused by Callosobruchus maculatus in cowpea during seed storage. Plant Arch 11(2):723–731
Di Matteo A, Bonivento D, Tsernoglou D, Federici L, Cervone F (2006) Polygalacturonase-inhibiting protein (PGIP) in plant defence: a structural view. Phytochemistry 67(6):528–533
Douglas AE (2018) Strategies for enhanced crop resistance to insect pests. Annu Rev Plant Biol 69:637–660
Dowling DK, Abiega KC, Arnqvist G (2007) Temperature-specific outcomes of cytoplasmic-nuclear interactions on egg-to-adult development time in seed beetles. Evolution 61(1):194–201
Downey MH, Searle R, Bellur S, Geiger A, Maitner BS, Ohm JR, Tuda M, Miller TE (2015) A comparative approach to testing hypotheses for the evolution of sex-biased dispersal in bean beetles. Ecol Evol 5(21):4819–4828
Downie D (2010) Baubles, bangles, and biotypes: a critical review of the use and abuse of the biotype concept. J Insect Sci 10(1):176
de los Reyes BG, (2019) Genomic and epigenomic bases of transgressive segregation-new breeding paradigm for novel plant phenotypes. Plant Science 288
Ehlers J, Hall A (1997) Cowpea (Vigna unguiculata L. Walp.). Field Crops Research 53(1–3):187–204
Fox C, Messina F (2018) Evolution of larval competitiveness and associated life-history traits in response to host shifts in a seed beetle. J Evolut Biol 31(2):302–313
Fox C, Bush M, Roff D, Wallin W (2004) Evolutionary genetics of lifespan and mortality rates in two populations of the seed beetle, Callosobruchus maculatus. Heredity 92(3):170–181
Fox CW, Wagner JD, Cline S, Thomas FA, Messina FJ (2009) Genetic architecture underlying convergent evolution of egg-laying behavior in a seed-feeding beetle. Genetica 136(1):179–187
Gompert Z, Messina FJ (2016) Genomic evidence that resource-based trade-offs limit host-range expansion in a seed beetle. Evolution 70(6):1249–1264
Gompert Z, Brady M, Chalyavi F, Saley TC, Philbin CS, Tucker MJ, Forister ML, Lucas LK (2019) Genomic evidence of genetic variation with pleiotropic effects on caterpillar fitness and plant traits in a model legume. Mol Ecol 28(12):2967–2985
Grazziotin MA, Cabral GB, Ibrahim AB, Machado RB, Aragão FJ (2020) Expression of the Arcelin 1 gene from Phaseolus vulgaris L. in cowpea seeds (Vigna unguiculata L.) confers bruchid resistance. Annals of Applied Biology 176(3):268–274
Guan Y, Stephens M (2011) Bayesian variable selection regression for genome-wide association studies and other large-scale problems. Ann Appl Stat 5(3):1780–1815
Haeger W, Henning J, Heckel DG, Pauchet Y, Kirsch R (2020) Direct evidence for a new mode of plant defense against insects via a novel polygalacturonase-inhibiting protein expression strategy. J Biol Chem 295(33):11833–11844
Horn LN, Shimelis H (2020) Production constraints and breeding approaches for cowpea improvement for drought prone agro-ecologies in Sub-Saharan Africa. Ann Agric Sci 65:83–91
Huang BE, Verbyla KL, Verbyla AP, Raghavan C, Singh VK, Gaur P, Leung H, Varshney RK, Cavanagh CR (2015) MAGIC populations in crops: current status and future prospects. Theor Appl Genet 128(6):999–1017
Huynh BL, Close TJ, Roberts PA, Hu Z, Wanamaker S, Lucas MR, Chiulele R, Cissé N, David A, Hearne S et al (2013) Gene pools and the genetic architecture of domesticated cowpea. Plant Genome 6(3):03
Huynh BL, Ehlers JD, Ndeve A, Wanamaker S, Lucas MR, Close TJ, Roberts PA (2015) Genetic mapping and legume synteny of aphid resistance in African cowpea grown in California. Mol Breed 35(1):1–9
Huynh BL, Matthews WC, Ehlers JD, Lucas MR, Santos JR, Ndeve A, Close TJ, Roberts PA (2016) A major QTL corresponding to the Rk locus for resistance to root-knot nematodes in cowpea. Theor Appl Gene 129(1):87–95
Huynh BL, Ehlers JD, Huang BE, Muñoz-Amatriaín M, Lonardi S, Santos JR, Ndeve A, Batieno BJ, Boukar O, Cisse N et al (2018) A multi-parent advanced generation inter-cross (MAGIC) population for genetic analysis and improvement of cowpea. Plant J 93(6):1129–1142
Kaewwongwal A, Chen J, Somta P, Kongjaimun A, Yimram T, Chen X, Srinives P (2017) Novel alleles of two tightly linked genes encoding polygalacturonase-inhibiting proteins (VrPGIP1 and VrPGIP2) associated with the br locus that confer bruchid (Callosobruchus spp.) resistance to mungbean (Vigna radiata) accession V2709. Frontiers in Plant Science 8:1692
Kaewwongwal A, Liu C, Somta P, Chen J, Tian J, Yuan X, Chen X (2020) A second VrPGIP1 allele is associated with bruchid resistance (Callosobruchus spp.) in wild mungbean accession acc41. Molecular Genetics and Genomics 295(2):275–286
Konrádhsdóttir H, Panagiotakopulu E, Lucas G (2021) A very curious larder-Insects from post-medieval Skálholt, Iceland, and their implications for interpreting activity areas. J Archaeol Sci 126:10531945
Kpoviessi D, Agbahoungba S, Agoyi EE, Chougourou D, Assogbadjo A (2019) Resistance of cowpea to cowpea bruchid (Callosobruchus maculatus Fab.): Knowledge level on the genetic advances. J Plant Breed Crop Sci 11(8):185–195
Langyintuo A, Lowenberg-DeBoer J, Faye M, Lambert D, Ibro G, Moussa B, Kergna A, Kushwaha S, Musa S, Ntoukam G (2003) Cowpea supply and demand in West and Central Africa. Field Crops Res 82(2–3):215–231
Liu HJ, Yan J (2019) Crop genome-wide association study: a harvest of biological relevance. Plant J 97(1):8–18
Lonardi S, Muñoz-Amatriaín M, Liang Q, Shu S, Wanamaker SI, Lo S, Tanskanen J, Schulman AH, Zhu T, Luo MC et al (2019a) The genome of cowpea (Vigna unguiculata [L.] Walp.). The Plant Journal 98(5):767–782
Lonardi S, Muñoz-Amatriaín M, Liang Q, Shu S, Wanamaker SI, Lo S, Tanskanen J, Schulman AH, Zhu T, Luo MC, Alhakami H, Ounit R, Hasan AM, Verdier J, Roberts PA, Santos JR, Ndeve A, Doležel J, Vrána J, Hokin SA, Farmer AD, Cannon SB, Close TJ (2019b) The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J 98(5):767–782
Lucas LK, Nice CC, Gompert Z (2018) Genetic constraints on wing pattern variation in Lycaeides butterflies: A case study on mapping complex, multifaceted traits in structured populations. Mol Ecol Resour 18(4):892–907
Lucas MR, Ehlers JD, Roberts PA, Close TJ (2012) Markers for quantitative inheritance of resistance to foliar thrips in cowpea. Crop Sci 52(5):2075–2081
Mano H, Toquenaga Y (2008) Wall-making behavior in Callosobruchus maculatus (Coleoptera: Bruchidae). Ann Entomol Soc Am 101(2):449–455
Messina FJ (1991) Life-history variation in a seed beetle: adult egg-laying vs larval competitive ability. Oecologia 85(3):447–455
Messina FJ (2004) Predictable modification of body size and competitive ability following a host shift by a seed beetle. Evolution 58(12):2788–2797
Messina FJ, Mitchell R (1989) Intraspecific variation in the egg-spacing behavior of the seed beetle Callosobruchus maculatus. J Insect Behav 2(6):727–742
Messina FJ, Jones JC, Mendenhall M, Muller A (2009) Genetic modification of host acceptance by a seed beetle, Callosobruchus maculatus (Coleoptera: Bruchidae). Ann Entomol Soc Am 102(1):181–188
Messina FJ, Lish AM, Gompert Z (2018) Variable responses to novel hosts by populations of the seed beetle Callosobruchus maculatus (Coleoptera: Chrysomelidae: Bruchinae). Environ Entomol 47(5):1194–1202
Messina FJ, Lish AM, Gompert Z (2019) Components of cowpea resistance to the seed beetle Callosobruchus maculatus (Coleoptera: Chrysomelidae: Bruchinae). J Econ Entomol 112(5):2418–2424
Messina FJ, Lish AM, Springer A, Gompert Z (2020) Colonization of marginal host plants by seed beetles (Coleoptera: Chrysomelidae): Effects of geographic source and genetic admixture. Environ Entomol 49:938–946
Miesho B, Hailay M, Msiska U, Bruno A, Malinga GM, Obia Ongom P, Edema R, Gibson P, Rubaihayo P, Kyamanywa S (2019) Identification of candidate genes associated with resistance to bruchid (Callosobruchus maculatus) in cowpea. Plant Breed 138(5):605–613
Mishra S, Macedo M, Panda S, Panigrahi J (2018) Bruchid pest management in pulses: past practices, present status and use of modern breeding tools for development of resistant varieties. Ann Appl Biol 172(1):4–19
Mishra SK, Nag A, Naik A, Rath SC, Gupta K, Gupta AK, Panigrahi J (2019) Characterization of host response to bruchids (Callosobruchus chinensis and C. maculatus) in 39 genotypes belongs to 12 Cajanus spp. and assessment of molecular diversity inter se. J Stored Prod Res 81:76–90
Mitchell R (1975) The evolution of oviposition tactics in the bean weevil, Callosobruchus maculatus (F). Ecology 56(3):696–702
Mitchell R (1990) Behavioral ecology of Callosobruchus maculatus. In: Bruchids and legumes: economics, ecology and coevolution, Springer, pp 317–330
Mitchell R (1991) The traits of a biotype of Callosobruchus maculatus (F)(Coleoptera: Bruchidae) from South India. J Stored Prod Res 27(4):221–224
Muchero W, Diop NN, Bhat PR, Fenton RD, Wanamaker S, Pottorff M, Hearne S, Cisse N, Fatokun C, Ehlers JD et al (2009) A consensus genetic map of cowpea [Vigna unguiculata (L) Walp.] and synteny based on EST-derived snps. Proceedings of the National Academy of Sciences 106(43):18159–18164
Muchero W, Roberts PA, Diop NN, Drabo I, Cisse N, Close TJ, Muranaka S, Boukar O, Ehlers JD (2013) Genetic architecture of delayed senescence, biomass, and grain yield under drought stress in cowpea. PloS ONE 8(7):e7004167
Munoz-Amatriain M, Sassoum L, Herniter I, Guo Y, Roberts P, Timothy J (2016) Development and characterization of a mini-core collection of cowpea. In: Conference report, American Society of Agronomy, pp 6–9
Muñoz-Amatriaín M, Mirebrahim H, Xu P, Wanamaker SI, Luo M, Alhakami H, Alpert M, Atokple I, Batieno BJ, Boukar O et al (2017) Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J 89(5):1042–1054
Nosil P, Villoutreix R, de Carvalho CF, Feder JL, Parchman TL, Gompert Z (2020) Ecology shapes epistasis in a genotype-phenotype-fitness map for stick insect colour. Nat Ecol Evol 4(12):1673–1684
Oas SE, D’Andrea AC, Watson DJ (2015) 10,000 year history of plant use at Bosumpra Cave, Ghana. Vegetation History and Archaeobotany. Ghana Veg Hist Archaeobot 24(5):635–653
Rollar S, Serfling A, Geyer M, Hartl L, Mohler V, Ordon F (2021) QTL mapping of adult plant and seedling resistance to leaf rust (Puccinia triticina Eriks.) in a multiparent advanced generation intercross (MAGIC) wheat population. Theor Appl Gene 134:37–51
Sanchez L, Farber C, Lei J, Zhu-Salzman K, Kurouski D (2019) Noninvasive and nondestructive detection of cowpea bruchid within cowpea seeds with a hand-held Raman spectrometer. Anal Chem 91(3):1733–1737
Schafleitner R, Sm Huang, Chu Sh, Jy Yen, Cy Lin, Mr Yan, Krishnan B, Ms Liu, Hf Lo, Cy Chen et al (2016) Identification of single nucleotide polymorphism markers associated with resistance to bruchids (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) and cultivated V. radiata through genotyping by sequencing and quantitative trait locus analysis. BMC Plant Biol 16(1):159
Schielzeth H, Rios Villamil A, Burri R (2018) Success and failure in replication of genotype-phenotype associations: How does replication help in understanding the genetic basis of phenotypic variation in outbred populations? Mol Ecol Res 18(4):739–754
Semeao AA, Campbell JF, Beeman RW, Lorenzen MD, Whitworth RJ, Sloderbeck PE (2012) Genetic structure of Tribolium castaneum (Coleoptera: Tenebrionidae) populations in mills. Environ Entomol 41(1):188–199
Solleti SK, Bakshi S, Purkayastha J, Panda SK (1841) Sahoo L (2008) Transgenic cowpea (Vigna unguiculata) seeds expressing a bean \(\alpha \)-amylase inhibitor 1 confer resistance to storage pests, bruchid beetles. Plant Cell Rep 27(12):1841-1850
Somta P, Jomsangawong A, Yundaeng C, Yuan X, Chen J, Tomooka N, Chen X (2018) Genetic dissection of azuki bean weevil (Callosobruchus chinensis L.) resistance in moth bean (Vigna aconitifolia [Jaqc.] Maréchal). Genes 9(11):555
Souframanien J, Gupta S, Gopalakrishna T (2010) Identification of quantitative trait loci for bruchid (Callosobruchus maculatus) resistance in black gram [Vigna mungo (L.) Hepper]. Euphytica 176(3):349–356
Southgate B (1979) Biology of the Bruchidae. Annu Rev Entomol 24(1):449–473
Soyk S, Benoit M, Lippman ZB (2020) New horizons for dissecting epistasis in crop quantitative trait variation. Annu Rev Gene 54:287–307
Storey JD, Bass AJ, Dabney A, Robinson D (2017) qvalue: Q-value estimation for false discovery rate control. http://github.com/StoreyLab/qvalue, R package version 2.15.0
Subramanyam B, Hagstrum D (2000) Botanicals. Alternatives to pesticides in stored-product IPM. Kluwer Academic Publishers, Boston, Massachussets, pp 303–320
Taggar GK, Arora R (2017) Insect biotypes and host plant resistance. In: Sandhu Surinder (ed) Breeding Insect Resistant Crops for Sustainable Agriculture. Springer, Nauni, pp 387–421
Tarver MR, Shade RE, Shukle RH, Moar WJ, Muir WM, Murdock LM, Pittendrigh BR (2007) Pyramiding of insecticidal compounds for control of the cowpea bruchid (Callosobruchus maculatus F). Pest Manag Sci 63(5):440–446
Thandar K, Laosatit K, Yimram T, Somta P (2021) Genetic analysis of seed resistance to Callosobruchus chinensis and Callosobruchus maculatus in cowpea. J Stored Prod Res 92:102
Toquenaga Y (1993) Contest and scramble competitions in Callosobruchus maculatus (Coleoptera: Bruchidae) II Larval competition and interference mechanisms. Popul Ecol 35(1):57–68
Toquenaga Y, Fujii K (1991) Contest and scramble competitions in Callosobruchus maculatus (Coleoptera: Bruchidae) I Larval competition curves and resource sharing patterns. Popul Ecol 33(2):199–211
Tuda M, Rönn J, Buranapanichpan S, Wasano N, Arnqvist G (2006) Evolutionary diversification of the bean beetle genus Callosobruchus (Coleoptera: Bruchidae): traits associated with stored-product pest status. Mol Ecol 15(12):3541–3551
Tuda M, Kagoshima K, Toquenaga Y, Arnqvist G (2014) Global genetic differentiation in a cosmopolitan pest of stored beans: effects of geography, host-plant usage and anthropogenic factors. PLoS One 9(9):e106268154
Weiss KM (2008) Tilting at quixotic trait loci (QTL): an evolutionary perspective on genetic causation. Genetics 179(4):1741–1756
Würschum T (2012) Mapping QTL for agronomic traits in breeding populations. Theor Appl Genet 125(2):201–210
Zhang Q, Yan Q, Yuan X, Lin Y, Chen J, Wu R, Xue C, Zhu Y, Chen X (2021) Two polygalacturonase-inhibiting proteins (VrPGIP) of Vigna radiata confer resistance to bruchids (Callosobruchus spp). J Plant Physiol 258:153376
Zhou X, Carbonetto P, Stephens M (2013) Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet 9(2):e1003264145
Acknowledgements
We thank Drs. T.J. Close, B.-L. Huynh, and P.A. Roberts at the University of California, Riverside, for valuable suggestions and for providing seeds of the MAGIC population. The support and resources from the Center for High Performance Computing at the University of Utah are gratefully acknowledged. C. Bovee, A. Hovinga, A. Israelsen, A. Springer and D. Weinerman provided technical assistance.
Funding
This work was funded by the Utah Agricultural Experiment Station (FJM, Paper Number 9429) and the National Science Foundation (ZG; DEB 1638768).
Author information
Authors and Affiliations
Contributions
FJM, AML and ZG designed the study, FJM and AML conducted the experiments, ZG analyzed the data, FJM and ZG wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Availability of data and material
Resistance data have been archived on Dryad (doi: https://doi.org/10.5061/dryad.x0k6djhjh).
Code availability
Perla and R scripts used for data analysis have been posted on GitHub (https://github.com/zgompert/CowpeaMapping.git).
Additional information
Communicated by Volker Hahn.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Messina, F.J., Lish, A.M. & Gompert, Z. Disparate genetic variants associated with distinct components of cowpea resistance to the seed beetle Callosobruchus maculatus. Theor Appl Genet 134, 2749–2766 (2021). https://doi.org/10.1007/s00122-021-03856-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03856-5