Abstract
Key message
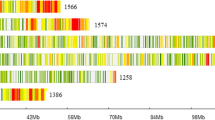
A SLAF-BSA approach was used to locate the RPF1 locus. The three most likely candidate genes were identified which provide a basic for cloning the resistance gene at the RPF1 locus.
Abstract
Spinach downy mildew is a globally devastating oomycete disease. The use of downy mildew resistance genes constitutes the most effective approach for disease management. Hence, the objective of the present study was to fine map the first-reported resistance locus RPF1. The resistance allele at this resistance locus was effective against races 1–7, 9, 11, 13, and 15 of Peronospora farinosa f. sp. spinaciae (P. effusa). The approach fine mapped RPF1 using specific-locus amplified fragment sequencing (SLAF-Seq) technology combined with bulked segregant analysis. A 1.72 Mb region localized on chromosome 3 was found to contain RPF1 based on association analysis. After screening recombinants with the SLAF markers within the region, the region was narrowed down to 0.89 Mb. Within this region, 14 R genes were identified based on the annotation information. To identify the genes involved in resistance, resequencing of two resistant inbred lines (12S2 and 12S3) and three susceptible inbred lines (12S1, 12S4, and 10S2) was performed. The three most likely candidate genes were identified via amino acid sequence analysis and conserved domain analysis between resistant and susceptible inbred lines. These included Spo12729, encoding a receptor-like protein, and Spo12784 and Spo12903, encoding a nucleotide-binding site and leucine-rich repeat domains. Additionally, based on the sequence variation in the three genes between the resistant and susceptible lines, molecular markers were developed for marker-assisted selection. The results could be valuable in cloning the RPF1 alleles and improving our understanding of the interaction between the host and pathogen.





Similar content being viewed by others
References
Baird NA, Etter PD, Atwood TS et al (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3:e3376
Correll JC, Morelock TE, Black M C et al (1994) Economically important diseases of spinach. Plant Dis 78:653–660
Cheng F, Sun R, Hou X et al (2016) Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat Genet 48:1218–1224
Correll JC, du Toit L, Koike ST et al (2010) Guidelines for spinach downy mildew: Peronospora farinosa f. sp. spinaciae (Pfs)
Correll JC, Bluhm BH, Feng C et al (2011) Spinach: better management of downy mildew and white rust through genomics. Eur J Plant pathol 129:193–205
Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826
Dardick C, Ronald P (2006) Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog 2:e2
Devran Z, Kahveci E, Özkaynak E et al (2015) Development of molecular markers tightly linked to Pvr4 gene in pepper using next-generation sequencing. Mol Breed 35(4):101
de Oliveira LFV, Christoff AP, de Lima JC et al (2014) The Wall-associated Kinase gene family in rice genomes. Plant Sci 229:181–192
Elshire RJ, Glaubitz JC, Sun Q et al (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379
Feechan A, Anderson C, Torregrosa L et al (2013) Genetic dissection of a TIR-NB-LRR locus from the wild North American grapevine species Muscadinia rotundifolia identifies paralogous genes conferring resistance to major fungal and oomycete pathogens in cultivated grapevine. Plant J 76:661–674
Feng C, Correll JC, Kammeijer KE et al (2014) Identification of new races and deviating strains of the spinach downy mildew pathogen Peronospora farinosa f. sp. spinaciae. Plant Dis 98:145–152
Feng C, Bluhm BH, Correll JC (2015) Construction of a spinach bacterial artificial chromosome (BAC) library as a resource for gene identification and marker development. Plant Mol Biol Rep 33:1996–2005
Geng X, Jiang C, Yang J et al (2016) Rapid identification of candidate genes for seed weight using the SLAF-Seq method in Brassica napus. PLoS ONE 11:e0147580
Graves H, Rayburn AL, Gonzalez-Hernandez JL et al (2016) Validating DNA polymorphisms using KASP assay in prairie cordgrass (Spartina pectinata Link) populations in the US. Front Plant Sci 6:1271
Hurni S, Scheuermann D, Krattinger SG et al (2015) The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc Natl Acad Sci 112:8780–8785
Irish BM, Correll JC, Koike ST et al (2003) Identification and cultivar reaction to three new races of the spinach downy mildew pathogen from the United States and Europe. Plant Dis 87:567–572
Irish BM, Correll JC, Feng C et al (2008) Characterization of a resistance locus (Pfs-1) to the spinach downy mildew pathogen (Peronospora farinosa f. sp. spinaciae) and development of a molecular marker linked to Pfs-1. Phytopathology 98:894–900
Jin W, Wang H, Li M et al (2016) The R2R3 MYB transcription factor PavMYB10. 1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol J 14:2120–2133
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664
Kubota M, Tamura O, Nomura Y et al (2017) Recent races of spinach downy mildew pathogen Peronospora farinosa f. sp. spinaciae in Japan. J Gen Plant Pathol 83:117–120
Liang D, Chen M, Qi X et al (2016) QTL mapping by SLAF-seq and expression analysis of candidate genes for aphid resistance in cucumber. Front Plant Sci 7:1000
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT methods. Methods 25:402–408
Martin GB, Brommonschenkel SH, Chunwongse J et al (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262:1432–1436
Meyers BC, Chin DB, Shen KA et al (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10:1817–1832
Meyers BC, Kaushik S, Nandety RS (2005) Evolving disease resistance genes. Curr Opin Plant Biol 82:129–134
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci 88:9828–9832
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4326
Nemri A, Atwell S, Tarone AM et al (2010) Genome-wide survey of Arabidopsis natural variation in downy mildew resistance using combined association and linkage mapping. Proc Natl Acad Sci 107:10302–10307
Plantum (2016) Denomination of Pfs: 16, a new race of downy mildew in spinach. http://www.plantum.nl/321519635/Document?documentregistrationid=780926983. Accessed 5 Sept 2016
Poulsen DME, Henry RJ, Johnston RP et al (1995) The use of bulk segregant analysis to identify a RAPD marker linked to leaf rust resistance in barley. Theor Appl Genet 91:270–273
Qian W, Feng CD, Zhang HL et al (2016) First report of race diversity of the spinach downy mildew pathogen, Peronospora effusa, in China. Plant Dis 100:1248
Qian W, Fan G, Liu D et al (2017) Construction of a high-density genetic map and the X/Y sex-determining gene mapping in spinach based on large-scale markers developed by specific-locus amplified fragment sequencing (SLAF-seq). BMC Genomics 18:276
Qin D, Dong J, Xu F et al (2015) Characterization and fine mapping of a novel barley Stage Green-Revertible Albino Gene (HvSGRA) by bulked segregant analysis based on SSR assay and specific length amplified fragment sequencing. BMC Genomics 16:838
Quarrie SA, Lazić-Jančić V, Kovačević D et al (1999) Bulk segregant analysis with molecular markers and its use for improving drought resistance in maize. J Exp Bot 50:1299–1306
Radwan O, Mouzeyar S, Nicolas P et al (2005) Induction of a sunflower CC-NBS-LRR resistance gene analogue during incompatible interaction with Plasmopara halstedii. J Exp Bot 56:567–575
She HB, Fan GY, Zhang HL et al (2017) Cloning and analysis of NBS-LRR type disease-resistant gene analogs in Spinach oleracea L. China Veg 5:26–33 (in Chinese)
Slusarenko AJ, Schlaich NL (2003) Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol Plant Pathol 4:159–170
Song WY, Wang GL, Chen LL et al (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804
Sun X, Liu D, Zhang X et al (2013) SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 8:e58700
Tör M, Brown D, Cooper A et al (2004) Arabidopsis downy mildew resistance gene RPP27 encodes a receptor-like protein similar to CLAVATA2 and tomato Cf-9. Plant Physiol 135:1100–1112
Venuprasad R, Dalid CO, Del Valle M et al (2009) Identification and characterization of large-effect quantitative trait loci for grain yield under lowland drought stress in rice using bulk-segregant analysis. Theor Appl Genet 120:177–190
Wang S, Cao J (2006) Advances in molecular mechanisms of the resistance of Arabidopsis thaliana to downy mildew in Chinese. Plant Prot 5:004
Wang Y, Tang H, DeBarry JD et al (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucl Acids Res 40:e49
Xia C, Chen L, Rong T et al (2015) Identification of a new maize inflorescence meristem mutant and association analysis using SLAF-seq method. Euphytica 202:35–44
Xiao S, Ellwood S, Calis O et al (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291:118–120
Xie W, Feng Q, Yu H et al (2010) Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc Natl Acad Sci 107:10578–10583
Xu C, Jiao C, Sun H et al (2017) Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat Commun 8:15275
Zheng W, Wang Y, Wang L et al (2016) Genetic mapping and molecular marker development for Pi65 (t), a novel broad-spectrum resistance gene to rice blast using next-generation sequencing. Theor Appl Genet 129:1035–1044
Acknowledgements
This work was performed at the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, Beijing, China, and was supported by National Natural Science Foundation of China (31401872), the National Science and Technology Pillar Program during the Twelfth Five-Year Plan Period of China (2014BAD01B08), Beijing Scientific Program of Municipal Commission of Science and Technology (Z171100001517014), the Chinese Academy of Agricultural Sciences Innovation Project (CAAS-ASTIP-IVFCAAS), and Fundamental Research Funds for Central Non-profit Scientific Institution (1610102018004).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Ethical standards
The authors declare that this study complies with the current laws of the countries in which the experiments were performed.
Additional information
Communicated by Richard G.F. Visser.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S6
Inoculation test. (a) Inoculation a seedling of the F2 population with Pfs9 using a sprayer. (b) Sporulation Pfs9 on a true leaf of an inoculated spinach seedling, representing a susceptible reaction to the Pfs9 (TIFF 20158 kb)
Fig. S7
Comparison of the amino acid sequences of the Spo12784 resistant and susceptible lines. Spo12784_R and Spo12784_S indicate the amino acid sequences of Spo12784 in the resistant and susceptible lines, respectively. The red arrow indicates that an amino acid mutation at the position results in a change of the conserved domain (TIFF 6252 kb)
Fig. S8
Comparison of the amino acid sequences of the Spo12903 resistant and susceptible lines. Spo12903_R and Spo12903_S indicate the amino acid sequences of Spo12903 in the resistant and susceptible lines, respectively. The red arrow indicates that an amino acid mutation at the position results in a change of the conserved domain (TIFF 7004 kb)
Fig. S9
Comparison of the amino acid sequences of the Spo12729 resistant and susceptible lines. Spo12729_R and Spo12729_S indicate the amino acid sequences of Spo12729 in the resistant and susceptible lines, respectively. The red arrow indicates that an amino acid mutation at the position results in change of conserved domain (TIFF 7105 kb)
Fig. S10
Comparison of the amino acid sequences of the Spo12762 resistant and susceptible lines. Spo12762_R and Spo12762_S indicate the amino acid sequences of Spo12762 in the resistant and susceptible lines, respectively (TIFF 5892 kb)
Fig. S11
Comparison of the amino acid sequences of the Spo12719 resistant and susceptible lines. Spo12719_R and Spo12719_S indicate the amino acid sequences of Spo12719 in resistant and susceptible lines, respectively (TIFF 3801 kb)
Fig. S12
Comparison of the amino acid sequences of the Spo12765 resistant and susceptible lines. Spo12765_R and Spo12765_S indicate the amino acid sequences of Spo12765 in resistant and susceptible lines, respectively (TIFF 7992 kb)
Fig. S13
Comparison of the conserved domain of seven genes (Spo12916, Spo12773, Spo12736, Spo12793, Spo12905, Spo12908, and Spo12730) in the resistant and susceptible lines (TIFF 1727 kb)
Rights and permissions
About this article
Cite this article
She, H., Qian, W., Zhang, H. et al. Fine mapping and candidate gene screening of the downy mildew resistance gene RPF1 in Spinach. Theor Appl Genet 131, 2529–2541 (2018). https://doi.org/10.1007/s00122-018-3169-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3169-4