Abstract
Key message
Rye genetic resources provide a valuable source of new alleles for the improvement of frost tolerance in rye breeding programs.
Abstract
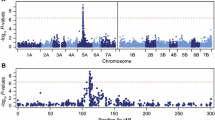
Frost tolerance is a must-have trait for winter cereal production in northern and continental cropping areas. Genetic resources should harbor promising alleles for the improvement of frost tolerance of winter rye elite lines. For frost tolerance breeding, the identification of quantitative trait loci (QTL) and the choice of optimum genome-based selection methods are essential. We identified genomic regions involved in frost tolerance of winter rye by QTL mapping in a biparental population derived from a highly frost tolerant selection from the Canadian cultivar Puma and the European elite line Lo157. Lines per se and their testcrosses were phenotyped in a controlled freeze test and in multi-location field trials in Russia and Canada. Three QTL on chromosomes 4R, 5R, and 7R were consistently detected across environments. The QTL on 5R is congruent with the genomic region harboring the Frost resistance locus 2 (Fr–2) in Triticeae. The Puma allele at the Fr–R2 locus was found to significantly increase frost tolerance. A comparison of predictive ability obtained from the QTL-based model with different whole-genome prediction models revealed that besides a few large, also small QTL effects contribute to the genomic variance of frost tolerance in rye. Genomic prediction models assigning a high weight to the Fr–R2 locus allow increasing the selection intensity for frost tolerance by genome-based pre-selection of promising candidates.



Similar content being viewed by others
References
Akar T, Francia E, Tondelli A, Rizza F, Stanca AM, Pecchioni N (2009) Marker-assisted characterization of frost tolerance in barley (Hordeum vulgare L.). Plant Breed 128:381–386
Båga M, Chodaparambil SV, Limin AE, Pecar M, Fowler DB, Chibbar RN (2007) Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct Integr Genom 7:53–68
Bauer E, Schmutzer T, Barilar I, Mascher M, Gundlach H, Martis MM, Twardziok SO, Hackauf B, Gordillo A, Wilde P, Schmidt M, Korzun V, Mayer KFX, Schmid K, Schön C-C, Scholz U (2017) Towards a whole-genome sequence for rye (Secale cereale L.). Plant J 89:853–869
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81:1084–1097
Browning BL, Browning SR (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84:210–223
Butler D, Cullis B, Gilmour A, Gogel B (2009) ASReml-R reference manual. Queensland Department of Primary Industries and Fisheries, Toowoomba
Campoli C, Matus-Cadiz MA, Pozniak CJ, Cattivelli L, Fowler DB (2009) Comparative expression of Cbf genes in the Triticeae under different acclimation induction temperatures. Mol Genet Genom 282:141–152
Case AJ, Skinner DZ, Garland-Campbell KA, Carter AH (2014) Freezing tolerance-associated quantitative trait loci in the Brundage × Coda wheat recombinant inbred line population. Crop Sci 54:982–992
Churchill G, Doerge R (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Collard BCY, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc B 363:557–572
Deng W, Nickle DC, Learn GH, Maust B, Mullins JI (2007) ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user’s datasets. Bioinformatics 23:2334–2336
Doležel J, Greilhuber J, Lucretti S, Meister A, Lysák MA, Nardi L, Obermayer R (1998) Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot 82:17–26
Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G (1998) Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor Appl Genet 97:968–975
Erath W, Bauer E, Kastirr U, Schmidt M, Korzun V, Schmiedchen B, Wilde P, Schön C-C (2016) Oligogenic control of resistance to soil-borne viruses SBCMV and WSSMV in rye (Secale cereale L.). Plant Breed 135:552–559
Fowler DB (2008) Cold acclimation threshold induction temperatures in cereals. Crop Sci 48:1147–1154
Fowler DB, Limin AE (1987) Exploitable genetic-variability for cold tolerance in commercially grown cereals. Can J Plant Sci 67:278
Fowler DB, Limin AE (2004) Interactions among factors regulating phenological development and acclimation rate determine low-temperature tolerance in wheat. Ann Bot 94:717–724
Fowler DB, Siminovitch D, Pomeroy MK (1973) Evaluation of an artificial test for frost hardiness in wheat. Can J Plant Sci 53:53–59
Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Tóth B, Hayes PM, Skinner JS, Pecchioni N (2004) Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter) × ‘Tremois’ (spring) barley map. Theor Appl Genet 108:670–680
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33:1–22
Galiba G, Vágújfalvi A, Li C, Soltész A, Dubcovsky J (2009) Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci 176:12–19
Geiger H, Miedaner T (2009) Rye breeding. In: Carena MJ (ed) Cereals. Springer, US, pp 157–181
Gray GR, Chauvin LP, Sarhan F, Huner N (1997) Cold acclimation and freezing tolerance (A complex interaction of light and temperature). Plant Physiol 114:467–474
Gusta LV, O’Connor BJ, MacHutcheon MG (1997) The selection of superior winter-hardy genotypes using a prolonged freeze test. Can J Plant Sci 77:15–21
Gusta LV, O’Connor BJ, Gao YP, Jana S (2001) A re-evaluation of controlled freeze-tests and controlled environment hardening conditions to estimate the winter survival potential of hardy winter wheats. Can J Plant Sci 81:241–246
Habier D, Fernando RL, Dekkers JCM (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177:2389–2397
Hackauf B, Rudd S, Van der Voort J, Miedaner T, Wehling P (2009) Comparative mapping of DNA sequences in rye (Secale cereale L.) in relation to the rice genome. Theor Appl Genet 118:371–384
Haldane J (1919) The combination of linkage values and the calculation of distances between the loci of linked factors. J Genet 8:299–309
Haseneyer G, Schmutzer T, Seidel M, Zhou RN, Mascher M, Schön C-C, Taudien S, Scholz U, Stein N, Mayer KFX, Bauer E (2011) From RNA-seq to large-scale genotyping-genomics resources for rye (Secale cereale L.). BMC Plant Biol 11:131
Haussmann B, Parzies H, Presterl T, Susic Z, Miedaner T (2004) Plant genetic resources in crop improvement. Plant Genet Resour 2:3–21
Holland J, Nyquist W, Cervantes-Martínez C (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22:9–112
Knox AK, Li CX, Vágújfalvi A, Galilba G, Stockinger EJ, Dubcovsky J (2008) Identification of candidate Cbf genes for the frost tolerance locus Fr-A m 2 in Triticum monococcum. Plant Mol Biol 67:257–270
Knox AK, Dhillon T, Cheng HM, Tondelli A, Pecchioni N, Stockinger EJ (2010) Cbf gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet 121:21–35
Korzun V, Malyshev S, Kartel N, Westermann T, Weber WE, Börner A (1998) A genetic linkage map of rye (Secale cereale L.). Theor Appl Genet 96:203–208
Li Y, Böck A, Haseneyer G, Korzun V, Wilde P, Schön C-C, Ankerst D, Bauer E (2011a) Association analysis of frost tolerance in rye using candidate genes and phenotypic data from controlled, semi-controlled, and field phenotyping platforms. BMC Plant Biol 11:146
Li Y, Haseneyer G, Schön C-C, Ankerst D, Korzun V, Wilde P, Bauer E (2011b) High levels of nucleotide diversity and fast decline of linkage disequilibrium in rye (Secale cereale L.) genes involved in frost response. BMC Plant Biol 11:6
Lu C-A, Ho TD, Ho S-L, Yu S-M (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. Plant Cell 14:1963–1980
Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang Z-X, Minobe Y, Yano M (2011) Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J 66:603–612
McCouch S, Baute GJ, Bradeen J, Bramel P, Bretting PK, Buckler E, Burke JM, Charest D, Cloutier S, Cole G, Dempewolf H, Dingkuhn M, Feuillet C, Gepts P, Grattapaglia D, Guarino L, Jackson S, Knapp S, Langridge P, Lawton-Rauh A, Lijua Q, Lusty C, Michael T, Myles S, Naito K, Nelson RL, Pontarollo R, Richards CM, Rieseberg L, Ross-Ibarra J, Rounsley S, Hamilton RS, Schurr U, Stein N, Tomooka N, van der Knaap E, van Tassel D, Toll J, Valls J, Varshney RK, Ward J, Waugh R, Wenzl P, Zamir D (2013) Agriculture: feeding the future. Nature 499:23–24
McIntosh R, Yamazaki Y, Dubcovsky J, Rogers W, Morris C, Appels R, Xia X (2013) Catalogue of Gene Symbols for Wheat. 12th International Wheat Genetic Symposium. Yokohama, Japan, September, 8–13, 2013
Meyer RS, Choi JY, Sanches M, Plessis A, Flowers JM, Amas J, Dorph K, Barretto A, Gross B, Fuller DQ, Bimpong IK, Ndjiondjop M-N, Hazzouri KM, Gregorio GB, Purugganan MD (2016) Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat Genet 48:1083–1088
Miedaner T (2013) Roggenanbau: Eine erfolgreiche Alternative. DLG-Verlag GmbH, AgrarPraxis kompakt
Mihaljevic R, Schön C-C, Utz HF, Melchinger AE (2005) Correlations and QTL correspondence between line per se and testcross performance for agronomic traits in four populations of European maize. Crop Sci 45:114–122
Millet EJ, Welcker C, Kruijer W, Negro S, Coupel-Ledru A, Nicolas SD, Laborde J, Bauland C, Praud S, Ranc N, Presterl T, Tuberosa R, Bedő Z, Draye X, Usadel B, Charcosset A, Van Eeuwijk F, Tardieu F (2016) Genome-wide analysis of yield in Europe: allelic effects vary with drought and heat scenarios. Plant Physiol 172:749–764
Möhring J, Piepho H-P (2009) Comparison of weighting in two-stage analysis of plant breeding trials. Crop Sci 49:1977–1988
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Pasquariello M, Barabaschi D, Himmelbach A, Steuernagel B, Ariyadasa R, Stein N, Gandolfi F, Tenedini E, Bernardis I, Tagliafico E, Pecchioni N, Francia E (2014) The barley Frost resistance-H2 locus. Funct Integr Genom 14:85–100
Petoukhov V, Semenov VA (2010) A link between reduced Barents-Kara sea ice and cold winter extremes over northern continents. J Geophys Res (Atmos) 115:D21111
Plaschke J, Börner A, Xie DX, Koebner RMD, Schlegel R, Gale MD (1993) RFLP mapping of genes affecting plant height and growth habit in rye. Theor Appl Genet 85:1049–1054
Pomeroy M, Fowler DB (1973) Use of lethal dose temperature estimates as indices of frost tolerance for wheat cold acclimated under natural and controlled environments. Can J Plant Sci 53:489–494
Schön C-C, Dhillon BS, Utz HF, Melchinger AE (2010) High congruency of QTL positions for heterosis of grain yield in three crosses of maize. Theor Appl Genet 120:321–332
Schwegler DD, Gowda M, Schulz B, Miedaner T, Liu W, Reif JC (2014) Genotypic correlations and QTL correspondence between line per se and testcross performance in sugar beet (Beta vulgaris L.) for the three agronomic traits beet yield, potassium content, and sodium content. Mol Breed 34:205–215
Shebeski L, McGinnis R, Evans L, Zuzens D (1973) Puma, a new cultivar of winter rye. Can J Plant Sci 53:67
Skinner DZ, Mackey B (2009) Freezing tolerance of winter wheat plants frozen in saturated soil. Field Crops Res 113:335–341
Skinner J, Zitzewitz J, Szűcs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger E, Thomashow M, Chen TH, Hayes P (2005) Structural, functional, and phylogenetic characterization of a large Cbf gene family in barley. Plant Mol Biol 59:533–551
Snape JW, Sarma R, Quarrie SA, Fish L, Galiba G, Sutka J (2001) Mapping genes for flowering time and frost tolerance in cereals using precise genetic stocks. Euphytica 120:309–315
Sorokina SA, Li C, Wettstein JJ, Kvamstø NG (2016) Observed atmospheric coupling between Barents Sea ice and the warm-Arctic cold-Siberian anomaly pattern. J Clim 29:495–511
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Tibshirani R (1996) Regression shrinkage and selection via the LASSO. J R Stat Soc Ser B (Methodol) 58:267–288
Tóth B, Francia E, Rizza F, Stanca AM, Galiba G, Pecchioni N (2004) Development of PCR-based markers on chromosome 5H for assisted selection of frost-tolerant genotypes in barley. Mol Breed 14:265–273
Utz HF (2011) PlabMQTL-Software for meta-QTL analysis with composite interval mapping. Version 0.9. Institute of Plant Breeding, Seed Science, and Population Genetics, University of Hohenheim
Van Ooijen JW (2006) JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma B V, Wageningen
Wimmer V, Albrecht T, Auinger H-J, Schön C-C (2012) synbreed: a framework for the analysis of genomic prediction data using R. Bioinformatics 28:2086–2087
Wimmer V, Lehermeier C, Albrecht T, Auinger H-J, Wang Y, Schön C-C (2013) Genome-wide prediction of traits with different genetic architecture through efficient variable selection. Genetics 195:573–587
Wooten DR, Livingston DP, Holland JB, Marshall DS, Murphy JP (2008) Quantitative trait loci and epistasis for crown freezing tolerance in the ‘Kanota’ × ‘Ogle’ hexaploid oat mapping population. Crop Sci 48:149–157
Würschum T, Longin CFH, Hahn V, Tucker MR, Leiser WL (2017) Copy number variations of Cbf genes at the Fr-A2 locus are essential components of winter hardiness in wheat. Plant J 89:764–773
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103:19581–19586
Zhao Y, Gowda M, Würschum T, Longin CFH, Korzun V, Kollers S, Schachschneider R, Zeng J, Fernando R, Dubcovsky J, Reif JC (2013) Dissecting the genetic architecture of frost tolerance in Central European winter wheat. J Exp Bot 64:4453–4460
Zhu J, Pearce S, Burke A, See D, Skinner D, Dubcovsky J, Garland-Campbell K (2014) Copy number and haplotype variation at the VRN-A1 and central FR-A2 loci are associated with frost tolerance in hexaploid wheat. Theor Appl Genet 127:1183–1197
Acknowledgements
This research was funded by the Federal Ministry of Education and Research (BMBF, Germany) within the project RYE SELECT (Grant ID 0315946). The authors are grateful to the field and lab team of KWS Lochow GmbH and to Stefan Schwertfirm and Amalie Fiedler from TUM for the technical assistance. The technical assistance of G. Schellhorn, Plant Sciences Department, University of Saskatchewan is also gratefully acknowledged. The authors thank Prof. Dr. H. Friedrich Utz (University of Hohenheim) for providing an extension of PlabMQTL which enabled the comparison of QTL and GP models.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical standards
The authors declare that this study complies with the current laws of the countries in which the experiments were performed.
Additional information
Communicated by Diane E. Mather.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Erath, W., Bauer, E., Fowler, D.B. et al. Exploring new alleles for frost tolerance in winter rye. Theor Appl Genet 130, 2151–2164 (2017). https://doi.org/10.1007/s00122-017-2948-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2948-7