Abstract
Key message
This study validated one QTL for adult plant resistance to stripe rust, identified donor lines of the resistance allele, and demonstrated that it is different from previously named Yr genes.
Abstract
The spread of more virulent and aggressive races of Puccinia striiformis f. sp. tritici (Pst, causal pathogen of stripe rust) after the year 2000 has caused substantial yield losses worldwide. To find new sources of resistance, we previously performed a genome-wide association study and identified a strong QTL for adult plant resistance on the short arm of chromosome 6B (QYr.ucw-6B). In this study, we validated QYr.ucw-6B in ten biparental populations, and mapped it 0.6 cM proximal to IWA7257 and 3.9 cM distal to IWA4408. We showed that QYr.ucw-6B is located approximately 15 cM proximal to the all-stage resistance gene Yr35 and that none of the resistant lines carries the previously cloned Yr36 gene. Based on these results, QYr.ucw-6B was assigned the name Yr78. This gene was not effective against Pst at the seedling stage, suggesting that it is an adult plant resistance gene. Yr78 has been effective against Pst races present in field experiments performed in the Western USA between 2011 and 2016. Since this gene is predicted to be present at low frequency in wheat germplasm from this region, it can provide a useful tool to diversify the sources of resistance against this devastating pathogen.
Similar content being viewed by others
Introduction
Stripe rust, caused by Puccinia striiformis Westend. f. sp. tritici Erikss. (Pst), is a devastating disease of wheat. More virulent and aggressive Pst races that began to appear around the year 2000 have spread rapidly over most of the wheat growing regions of the world, causing significant yield losses (Chen 2005; Hovmøller et al. 2008, 2016; Milus et al. 2008; Wellings 2011). These new Pst races are more aggressive, have broader virulence profiles, and are tolerant to higher temperatures than previously identified races (Markell and Milus 2008; Milus et al. 2008). A comparison with reference isolates sampled from six continents suggested that the post-2011 European races of Pst originated from the center of diversity of the pathogen in the near-Himalayan region of South Asia (Ali et al. 2014; Hovmøller et al. 2016).
While stripe rust can be controlled by the timely application of fungicides, control through the deployment of resistant cultivars is a more economically viable and environmentally friendly solution. Unfortunately, many of the previously characterized Pst resistance genes are not effective against these new Pst races, forcing wheat breeders to look for new sources of resistance. Pst all-stage resistance genes Yr5 (Chen et al. 2003) and Yr15 (Yaniv et al. 2014) have remained effective to these new races and have been combined into several wheat varieties (e.g. Patwin 515, Summit 515, Blanca Grande 515, etc.). However, the recent release of commercial varieties carrying each of these genes separately is jeopardizing previous gene stewardship strategies and making more urgent the identification of additional resistance genes.
Stripe rust resistance genes are broadly classified into all-stage and adult plant resistance (APR) genes. All-stage resistance genes, which are effective since early stages of plant development, are typically race-specific, and frequently encode NBS-LRR resistance proteins (Lowe et al. 2011). By contrast, APR genes are effective later in development and often, are effective to a broader range of races, confer partial resistance, and encode a more diverse set of proteins (Chen 2013; Fu et al. 2009; Krattinger et al. 2009; Lowe et al. 2011). To expand the list of APR genes against Pst in hexaploid wheat (Triticum aestivum L.), we previously performed a global genome-wide association study (GWAS) in six different environments in the western USA between the years 2011 and 2013 (Maccaferri et al. 2015).
In this GWAS, we identified ten loci that were highly significant in most of the environments and had a low experiment-wise error rate (Maccaferri et al. 2015). Among these loci, we identified one on chromosome 6B (QYr.ucw-6B) linked to SNP locus IWA7257 that was mapped in a different location from previously named Pst resistance genes. This locus was highly significant for APR in field trials (infection type P < 0.00001), but was not significant at the seedling stage under controlled environments for any of the races (PSTv-4, PSTv-14, PSTv-37, and PSTv-40) tested in the previous study (Maccaferri et al. 2015). This locus was also highly significant in a second GWAS performed in the same study in which accessions with highly resistant infection types (ITs of 0–3) were excluded from the analyses. The last result suggested that this locus confers partial resistance to Pst (Maccaferri et al. 2015).
Even though the previous GWAS provided valuable information about the chromosome locations of multiple QTL, an experimental validation is still required to identify which of the accessions carrying the favorable SNP allele actually carry the associated resistance gene. This validation is particularly important for wheat breeders to identify germplasm sources from which the resistance alleles can be introgressed into their breeding programs. The identification of correct donor lines is also critical to develop high-density genetic maps and, eventually, to clone the resistance gene. GWAS in wheat are usually insufficient to identify candidate genes because linkage disequilibrium in this species frequently extends over several cM due to its self-pollinating nature and recent origin (Chao et al. 2010; Maccaferri et al. 2015).
In this study, we report the validation of QYr.ucw-6B in multiple biparental populations and the identification of multiple donors of the resistance alleles that can be used as parental lines in wheat breeding programs. We also report the difference between this QTL and previously named Pst resistance genes on chromosome arm 6BS, which supports its official designation as Yr78 (QYr.ucw-6B). We show that this gene is more effective in adult plants, and propose that it will constitute a useful tool for wheat breeding programs interested in diversifying the sources of genetic resistance against the new virulent races of Pst.
Materials and methods
Development of biparental populations
From the GWAS described above (Maccaferri et al. 2015), we selected 10 accessions that carry the resistance allele for QYr.ucw-6B (Table 1). All of these accessions are publicly available in the United States Department of Agriculture (USDA) National Small Grains Collection (NSGC). Table 1 summarizes their accession numbers, country of origin, year received by the National Plant Germplasm System and alleles present in the markers at the peak of the QTL.
The putative donors of the resistance allele were crossed with the susceptible variety Avocet ‘S’, which has been used as a recurrent parent in the generation of near isogenic lines for many Yr genes (McIntosh et al. 1995; Wan and Chen 2014). The F1s were self-pollinated in the greenhouse and F2 populations were developed, including 94 individuals each. The first seven F2 populations were evaluated during the 2014–2015 growing season and F3 families derived from F2 plants homozygous for the peak marker were evaluated during the following growing season. Three additional F2 populations for QYr.ucw-6B were evaluated only in the 2015–2016 growing season. Plants were genotyped for IWA7257, the marker associated with the peak of QYr.ucw-6B.
All field trials were performed at the University of California Experimental Field Station in Davis, California (38°31′33″N, 121°46′30″W, elevation 16 m; henceforth UCD). The experiments were surrounded with a susceptible border (D6301) that was inoculated with Pst spores collected from the previous field season.
Genotyping of the biparental populations
Leaf tissues from individual F2 plants and their respective parental lines were collected at the five-leaf stage. Genomic DNA was extracted using previously published methods (Anderson et al. 1993). All the bi-parental populations were genotyped using a KASP assay (Kompetitive Allele Specific PCR, LGC-Genomics, UK) (Semagn et al. 2014). The targeted SNP was at the 3′ end of the primers, following standard KASP guidelines. The allele-specific primers for IWA7257 (QYr.ucw-6B) were IWA7257_Rev_A_VIC (GAAGGTCGGAGTCAACGGATTagaccctacgacgttagcga) and IWA7257_Rev_C_FAM (GAAGGTGACCAAGTTCATGCTagaccctacgacgttagcgc), with common primer IWA7257_Com1 (attggaatcagctgggtcat). The capital letters of the primers indicate the VIC and FAM tails and the 3′ allele-specific nucleotide is underlined. The length of the amplicon was 77 bp.
The primer assay mix (100 µl) included 12 µl VIC primer (100 mM), 12 µl FAM primer (100 mM), 30 µl common primer (100 mM), and 46 µl distilled water. KASP assays were performed in a 5.07 µl reaction volume (2.5 µl 2× KASP Master Mix, 0.07 µl KASP primer assay mix and 2.5 µl genomic DNA at 5–50 ng µl−1). A two-step touchdown PCR was carried out using the following conditions: 94 °C for 15 min, followed by ten cycles of touchdown of 94 °C for 20 s, annealing from 61 to 55 °C for 1 min (dropping 0.6 °C per cycle), followed by 26 cycles of 94 °C for 20 s, annealing at 55 °C for 1 min. KASP results were analyzed with a FLUOstar Omega F plate reader (BMG LABTECH, Ortenberg, Germany) using the software KlusterCaller (LGC Genomics, Teddington, UK).
For the PI 519805 × Avocet ‘S’ population we developed a complete genetic map to identify plants carrying only the resistance gene underlying QYr.ucw-6B. This population was genotyped using the Infinium wheat SNP 9 K iSelect assay (Illumina Inc., San Diego, CA, USA) (Cavanagh et al. 2013) at the USDA-ARS genotyping laboratory at Fargo, North Dakota. A total of 2821 polymorphic SNP markers were obtained and used for genetic map construction. The map of the 6B chromosome was supplemented with six polymorphic simple sequence repeats (SSRs, GrainGenes database http://wheat.pw.usda.gov/GG3/).
Field inoculation and evaluation of stripe rust severity
All populations were sown in the field in mid-November surrounded by Pst susceptible borders and interspersed with susceptible plants to facilitate the spread of the disease. Susceptible spreader lines were also planted between every population. The F2 populations were evaluated as single plants in 1-m rows including five plants per row, with a separation of 30 cm between rows to facilitate disease evaluation. Individual F2 plants were genotyped with IWA7257. Selected F3 families derived from F2 lines homozygous for IWA7257 were evaluated in rows with at least 30 plants each in the 2015–2016 growing season.
The field was inoculated in February with a mixture of Pst spores from the previous year. The Pst races collected from infected wheat plants at the UC Davis field in 2015 and 2016, and their virulence formulas are described in Table 2. During both growing seasons, all susceptible spreader rows and borders became fully infected with Pst, providing a strong and uniform disease pressure.
Wheat lines were evaluated for Pst resistance twice, approximately two weeks before and two weeks after heading, to minimize potential escapes, and to avoid the death of plants severely affected by the disease before the scoring. The second scoring date occurred during heavier infection and was preferred for the statistical analysis. Infection type (IT) was scored using the McNeal’s 0 (resistant)–9 (susceptible) scale (Line and Qayoum 1992). Disease severity (SEV) was scored as the percentage of leaf area infected.
Evaluation of seedling resistance to Pst
To test the effect of QYr.ucw-6B on seedling resistance, we selected four BC1F2 plants derived from the cross PI 519805/2*Avocet ‘S’ that were homozygous for the QYr.ucw-6B resistance allele, and homozygous for the susceptible allele for the other resistance QTL detected in this population, and inoculated them with Pst spores collected in the field in 2015. As positive control, we used a line homozygous for Yr35 that was generously provided by Professor Robert Park (Plant Breeding Institute, University of Sydney). In addition, we evaluated seedlings from two F3 families homozygous for the QYr.ucw-6B resistance allele, three homozygous for the susceptibility allele, one line homozygous for the all-stage resistance gene Yr35, and the susceptible control Avocet ‘S’ with races PSTv-4, PSTv-14, PSTv-37, PSTv-40 and PSTv-51 at Washington State University.
Pst reactions at the seedling stage were evaluated in a CONVIRON growth chambers (PGR15) using protocols similar to those described before (Chen et al. 2002; Pahalawatta and Chen 2005). Briefly, plants were inoculated at the two-leaf stage using spores collected in the field in 2015, and were kept in the dark at 90% humidity and 10 °C for 48 h. Plants were then transferred into a diurnal temperature cycle that changed gradually from 10 to 20 °C with 16 h photoperiod. The chambers had metal halide and high-pressure sodium light that provided a light intensity of ~260 µM m−2 s−1. Infection types (ITs) were recorded 20–22 days after inoculation using the McNeal’s 0–9 scale (Line and Qayoum 1992).
Statistical and QTL analyses
For each population segregating for QYr.ucw-6B, the genotype at the marker IWA7257 was used as a class variable in one-way ANOVAs for infection type and severity. Normality of residuals was tested using the Shapiro–Wilk test and homogeneity of variances using Levene’s tests as implemented in SAS 9.4 (SAS Institute Inc., Cary, NC, USA). For those populations that did not meet these assumptions, we first tested different power transformations, and if they still did not meet the assumptions, we analyzed those populations using a non-parametric test (Kruskal–Wallis, as implemented in SAS 9.4).
For the population PI519805 × Avocet ‘S’ segregating for IWA7257, a genetic map was constructed with JoinMap 4.0 (Van Ooijen 2006) and MAPMAKER/EXP 3.0 (Lincoln et al. 1992) using the Kosambi mapping function (Kosambi 1944). A minimum LOD (logarithm of odds) threshold of 2.0 was used for grouping markers into linkage groups, and a three-point linkage analysis was carried out to determine the most likely order of linked markers. Linkage groups were assigned to chromosomes using a previous consensus map (Cavanagh et al. 2013). Completely linked markers were merged and only one of the markers for each group was used for the QTL analysis. For groups of linked markers known to be in the same chromosome but for which no significant linkage was detected between groups, the distances were estimated from the consensus map (Cavanagh et al. 2013). Those estimated distances are indicated in parenthesis and marked with an “*” in the maps.
QTL analyses were performed with Windows QTL Cartographer V2.5 (Wang et al. 2012) using composite interval mapping (CIM) with both backward and forward regressions at α = 0.1. A threshold (LOD) value of 3.0 was used to determine the presence of significant QTL.
Results
Validation populations for QYr.ucw-6B (Yr78)
The ten accessions selected for Yr78 showed good adult plant resistance in the field (supplementary Fig. S1 A–J). In all ten segregating populations derived from these accessions, we detected significant effects associated with SNP marker IWA7257 (peak marker of QYr.ucw-6B in GWAS) for both Pst infection type and severity (Table 1). This result suggests that IWA7257 is a relatively good predictor of QYr.ucw-6B. The association between IWA7257 and Pst resistance was further validated during the 2015–2016 growing season using F3 families derived from homozygous F2 plants. All seven F3 populations showed highly significant differences in Pst infection type and severity between families homozygous for the different IWA7257 alleles (Table 3).
On average, the IWA7257 marker explained 21.4 ± 3.9% of the variation in IT and 19.9 ± 3.9% of the variation in severity in the F2 experiment (Table 1) and 47.4 ± 6.3% of the variation in IT and 44.5% ± 5.9% of the variation in severity in the F3 experiment (Table 3). In both the F2 and F3 experiments, the population generated from the cross PI 519805 × Avocet ‘S’ showed the largest proportion of variation explained by the IWA7257 marker (Tables 1, 3), and was selected for the construction of a complete linkage map, and for the identification of lines segregating only for QYr.ucw-6B.
PI 519805 × Avocet ‘S’ linkage map
We generated a complete linkage map of the PI 519805 × Avocet ‘S’ population to evaluate the presence of other Pst resistance QTL. This information was used to select plants segregating only for QYr.ucw-6B for further evaluation with different Pst races. We genotyped the complete F2 population using the Illumina iSelect 9K SNP assay (Cavanagh et al. 2013), six SSR markers from chromosome 6B, two from chromosome 6D (Somers et al. 2004), and marker csLV46 (Dr. Evans S Lagudah, personal communication). Marker csLV46 was previously mapped linked to Yr29 on chromosome arm 1BL (Kolmer et al. 2015; Lan et al. 2014). A total of 2821 polymorphic SNP markers were identified, and mapped to 689 unique loci covering 2425 cM. Linkage groups were assigned to chromosomes based on previously published consensus SNP maps (Cavanagh et al. 2013). Fewer SNP markers were mapped on chromosomes from the D genome (221 markers) than on those from the A (1072 markers) or B (1491 markers) genomes (37 markers were ungrouped). A lower proportion of markers in the D genome was also reported for the SNP consensus map, which was generated with the same SNP assay (Cavanagh et al. 2013).
Markers from chromosomes 5B and 7B were linked in this population suggesting that PI 519805 (ND 457*3/T.durum//Estanzuela Dakuro) carries the centromeric reciprocal translocation 5BL-7BL and 5BS-7BS, which was first identified in T. aestivum ssp. compactum (Sears 1953) and was prevalent in West European wheats in the 1960s and 1970s and in their descendants (Riley et al. 1967). The short and long arms of the 5B and 7B maps were manually separated and presented without the translocation in supplementary Fig. S2.
In this population, IWA7257 was mapped on the short arm of chromosome 6B, 0.6 cM proximal to SSR marker wmc494 and 4.5 cM distal to linked markers IWA4408 (SNP) and wmc737 (SSR) (Fig. 1a). This differs from the position of IWA7257 on the long arm of chromosome 6B (scaled position 112.3 cM) in the 9K iSelect assay SNP consensus map (Cavanagh et al. 2013). A comparison of the 200 bp sequence flanking SNP IWA7257 with the IWGSC draft sequence of the different chromosome arms of Chinese Spring (https://urgi.versailles.inra.fr/blast/blast.php) showed 100% identity to sequences from chromosome arm 6BS (BLASTN E = 3e−98) and no significant similarity to 6BL, which confirmed that IWA7257 is located on the short arm of chromosome 6B.
Comparative maps of QYr.ucw-6B and Yr35. a Linkage map and QTL analysis based on F2 plants from the population PI 519805 × Avocet ‘S’. b Linkage map of the population PI 494101 × Avocet ‘S’. The position of QYr.ucw-6B as a Mendelian gene in the two linkage maps is based on F3 field data. c Map of Pst all-stage resistance gene Yr35 based on Dadkhodaie et al. (2011) including common marker gwm508. d Evaluation of the presence of Yr36 in the ten lines confirmed to carry QYr.ucw-6B. DIC = T. turgidum ssp. dicoccoides carrying the Yr36 gene (positive control)
PI 519805 × Avocet ‘S’ QTL analysis
Using a threshold LOD score of 3.0, we identified two significant QTL for infection type and severity on chromosome arms 6BS (QYr.ucw-6B, Fig. 1a) and 1BL (QYr.ucw-1B, supplementary Fig. S3). In a factorial ANOVA including both QTL, QYr.ucw-6B explained 45.9% of the variation in IT and 49.7% of the variation in severity (slightly different from the values in Table 1 calculated from one-way ANOVAs), whereas QYr.ucw-1B explained 30% of the variation in IT and 27% of the variation in severity. No significant interaction was detected between these two QTL suggesting that their contributions to Pst resistance are mainly additive (independent from each other).
The peak of QYr.ucw-6B (based on F2 data), was mapped between markers IWA7257 and wmc737/IWA4408 on chromosome arm 6BS (Fig. 1a). This location was confirmed using data from F3 families homozygous for the resistance and susceptibility alleles. In population PI 519805 × Avocet ‘S’, QYr.ucw-6B was mapped 0.6 cM proximal to IWA7257 and 3.9 cM distal to wmc737/IWA4408 (Fig. 1a). Although a similar location was detected for population PI 494101 × Avocet ‘S’ (Fig. 1b), the position of the QTL in the PI 519805 × Avocet ‘S’ map is probably more precise because we had information of the additional resistance allele segregating in this population. In summary, the results of the three mapping experiments consistently indicate that QYr.ucw-6B is located within the IWA7257-wmc737 interval. We analyzed each of the critical recombination events and provided our best estimate of the position of the underlying Yr78 gene within this interval (Fig. 1a, b). However, the results presented here should be considered only as a first step in the Mendelization of QYr.ucw-6B, because these populations were not generated from isogenic lines, and segregation for other Pst resistance genes in the background can generate some imprecisions in our mapping results.
The QTL mapped on the distal region of chromosome arm 1BL (peak at SNP marker IWA802, LOD = 5.7) was designated as QYr.ucw-1B based on its similar position to a QTL mapped in a previous GWAS (Maccaferri et al. 2015). IWA802 was mapped 3 cM proximal to csLV46 (supplementary Fig. S3), a marker that has been previously mapped linked to the leaf rust/stripe rust APR locus Lr46/Yr29 (Kolmer et al. 2015). Based on the comparison with the map published by Kolmer et al. (2015), it is likely that QYr.ucw-1B corresponds to Yr29 (supplementary Fig. S3).
Comparison of QYr.ucw-6B with other Pst resistance genes on chromosome arm 6BS
To determine if QYr.ucw‐6B was different from Pst resistance genes Yr35 and Yr36 previously mapped on the same chromosome arm, we compared their positions on the 6BS map and their response to different Pst races at the seedling stage.
The all-stage resistance gene Yr35 was previously mapped between SSR markers gwm191 and cfd1 (Dadkhodaie et al. 2011). Although we were unable to map these two SSR markers in our segregating populations, we were able to map gwm508 (the next SSR marker proximal to Yr35) in the population PI 494101 × Avocet ‘S’ (Fig. 1b). Yr35 was mapped 7.5 cM distal to gwm508, whereas IWA7257 was mapped 7 cM proximal to gwm508 (Fig. 1c). Since QYr.ucw‐6B was mapped proximal to IWA7257, the previous results indicate that QYr.ucw‐6B is at least 15 cM proximal to Yr35 (compare Fig. 1b and c), and that they are different genes. This hypothesis is also supported by different reactions to Pst inoculations at the seedling stage described in the next section.
The Yr36 gene was previously cloned and was shown to encode a protein carrying a START and a kinase domain, designated WKS1 (Fu et al. 2009). We used primers specific for the WKS1 START domain (Fu et al. 2009) to test for the presence of this gene in the ten populations segregating for Yr78. We amplified the expected 871- and 537-bp PCR fragments from the positive control line T. turgidum ssp. dicoccoides accession FA15-3 (DIC, Fig. 1d). The 871-bp fragment corresponds to WKS1 and the 537-bp to the closely linked non-functional paralog WKS2 (Gou et al. 2015). The ten lines carrying the QYr.ucw-6B resistance allele (Table 1) did not show any PCR amplification product using the same set of primers (Fig. 1d). This result confirmed that Yr36 is not present in these lines and that, therefore, QYr.ucw-6B and Yr36 are different genes. Based on the Yr35 and Yr36 results, QYr.ucw-6B was assigned the official name Yr78.
Seedling reaction for QYr.ucw-6B
The ten lines validated for the presence of Yr78 (Table 1) showed relatively high levels of resistance to Pst in the field at the adult plant stage (supplementary Fig. S1, A–J). At the seedling stage, all BC1F2 plants from the cross PI 519805/2*Avocet ‘S’ homozygous for Yr78 were as susceptible to Pst (field collected spores, supplementary Fig. S1 K) as the susceptible control Avocet ‘S’ (supplementary Fig. S1 K). By contrast, seedlings homozygous for Yr35 (Dadkhodaie et al. 2011) grown in the same chamber showed a clear resistance reaction to the same Pst spores (supplementary Fig. S1 K).
Similar results were obtained for F3 seedlings from the PI519805 × Avocet ‘S’ cross inoculated with Pst races PSTv-4, PSTv-14, PSTv-37, PSTv-40, and PSTv-51 in a growth chamber (Table 4). F3 seedlings homozygous for Yr78 and those homozygous for the susceptibility allele, both showed high levels of infection type (mainly ITs 7 and 8), similar to those observed in the susceptible recurrent parent Avocet ‘S’ (Table 4). As in the experiment using Pst urediniospores collected in the field, the seedlings carrying Yr35 showed high levels of resistance to the five individual races (mainly ITs 1–3). These results confirmed that Yr35 and Yr78 are different genes and that Yr78 is an APR gene.
Discussion
New Pst races that appeared around the year 2000 in the USA and later in Australia, Europe and North Africa have a broader virulence profile, increased aggressiveness and better tolerance to high temperatures than previous races. These races have defeated many of the previously known stripe rust resistance genes, depleting the options available to wheat breeders. To expand the set of resistance genes we performed a GWAS and identified several potential new genes (Maccaferri et al. 2015). In this study, we validated and characterized one of these resistance genes on wheat chromosome arm 6BS (Yr78), and showed that it is different from stripe rust resistance genes previously identified in this chromosome arm.
Relation between Yr78 and other resistance genes and QTL mapped to chromosome arm 6BS
New official Yr names are assigned to Pst resistance loci that are different from previously named Yr genes, and for which donor lines are available in public seed repositories. QYr.ucw‐6B satisfied both requirements. The donor lines of this Pst resistance QTL are all deposited in the USDA National Small Grains Collection, and are available upon request. In addition, we showed that Yr78 is different from Pst resistance genes Yr35 and Yr36, previously mapped on chromosome arm 6BS.
Using a common SSR marker, we mapped Yr78 roughly 15 cM proximal to the known location of Yr35 (Fig. 1), and showed that, at the seedling stage, these two genes differ in their reactions to multiple Pst races. Seedlings carrying Yr35 were resistant to all tested Pst races, whereas seedlings carrying Yr78 showed infection types and severities similar to those observed in the susceptible parent Avocet ‘S’ (supplementary Fig. S1). Finally, Yr35 was transferred to hexaploid wheat from T. turgidum ssp. dicoccoides (Marais et al. 2005), several years after the development of the cultivars carrying QYr.ucw-6B reported in this study (Table 1). Taken together, the previous arguments consistently support the hypothesis that Yr78 and Yr35 are different genes.
We also demonstrated that QYr.ucw-6B is different from the previously cloned resistance gene Yr36 (Fu et al. 2009) since primers diagnostic for Yr36 failed to amplify any products in the lines validated for QYr.ucw-6B in this study (Fig. 1d). Based on the previous results QYr.ucw-6B was officially assigned the name Yr78.
We also explored QTL previously mapped close to the Yr78 region to determine if they are synonymous of the new designated Yr gene. Of the five QTL previously mapped close to the region including Yr78 on chromosome 6B, three were mapped on the long arm, and are likely different genes. These include QYr.caas-6B (Lan et al. 2010), QYr-6B (William et al. 2006), and QYr.inra-6B (Dedryver et al. 2009). By contrast, QYr.sun-6B identified in the Australian variety Janz (Bariana et al. 2010) and QYr.wgp-6BS.1 identified in the US wheat variety Stephens (Santra et al. 2008) were both mapped on the 6BS arm distal to gwm644, within regions that overlap with Yr78 (Fig. 1a). Janz and Stephens, both carry the “T” allele at IWA7257 (=AA), which accurately predicted the presence of Yr78 in the ten populations validated in this study. Based on these results, we propose that both Janz and Stephens carry Yr78, and suggest that QYr.sun-6B and QYr.wgp-6BS.1 be designated as synonymous of Yr78. To the best of our knowledge, QYr.wgp-6BS.1 should be credited as the first report for Yr78 (Santra et al. 2008).
IWA7257, the closest marker to Yr78, was also reported to be significantly associated with Pst resistance in two GWAS conducted in the Pacific North West (PNW). The first one, used a core subset of 1175 accessions from the NSGC winter wheat germplasm collection (Bulli et al. 2016), and the other one used 402 advanced winter wheat accessions from the PNW winter panel (Naruoka et al. 2015). These results suggest that Yr78 is present in both spring and winter wheat cultivars.
All-stage versus partial adult plant resistance genes
In the two GWAS for winter wheat (Bulli et al. 2016; Naruoka et al. 2015), as well as in the GWAS for spring wheat (Maccaferri et al. 2015), the IWA7257 marker was associated with APR to Pst in the field, but not with seedling resistance in inoculations performed in controlled environment experiments (Table 4; supplementary Fig. S1). In this study, we confirmed that Yr78 is not effective at the seedling stage, but that it confers partial resistance to Pst at the adult plant stage (supplementary Fig. S1).
The classification of Yr78 as a partial APR gene has some practical implications. First, the few cloned wheat genes conferring partial resistance to Pst at the adult plant stage, have revealed protein architectures different from the canonical NBS-LRR resistance genes (Fu et al. 2009; Krattinger et al. 2009). The identification of such genes provides the opportunity to diversify the resistance mechanisms used to control this rapidly evolving pathogen. In addition, some of these partial APR genes have been historically more durable than many of the all-stage resistance genes, which can be rapidly defeated by changes or deletions of the recognized effectors (Lowe et al. 2011). The classification of Yr78 as a partial APR gene will likely increase the interest in cloning this gene.
Introgression of Yr78 into breeding programs
The “T” (=AA) allele of IWA7257 was predictive of the presence of the resistance gene in all ten populations selected for validation in this study. This allele was also predictive of the presence of a Pst resistance QTL on 6BS (likely Yr78) in the varieties Janz (Bariana et al. 2010) and Stephens (Santra et al. 2008). These results suggest that IWA7257 is genetically close to Yr78 and that limited historical recombination has occurred between this marker and the resistance gene. Even though the “T” allele of IWA7257 has been a good predictor of the presence of Yr78, we have found some putative recombination events between IWA7257 and Yr78 in our segregating populations. This suggests that IWA7257 is not a perfect marker for Yr78, and that it should be used cautiously to predict the presence of Yr78 in uncharacterized germplasm.
Among the winter wheat varieties, the frequency of the IWA7257 “T” allele associated with Pst resistance was 24% in the NSGC winter wheat core collection (1175 genotyped accessions) (Bulli et al. 2016), and 46% in the PNW winter panel (Naruoka et al. 2015). Among the spring wheats in the NSGC core collections, the IWA7257 “T” allele was present on average in 23% of the hexaploid wheat accessions and 1.3% of the tetraploid accessions. Therefore, selection for Yr78 can be used to improve Pst resistance in a large number of lines in both tetraploid and hexaploid wheat breeding programs and to diversify the Pst resistance genes currently used to control this pathogen.
So far, the effectiveness of Yr78 against the new Pst races has been demonstrated only in the western USA and Australia (Bariana et al. 2010). Therefore, before deploying this gene in other regions, it will be necessary to confirm its effectiveness against the local Pst races. If effective, Yr78 will particularly useful in Asia, where the favorable IWA7257 allele is present at low frequency (<10%) (Maccaferri et al. 2015).
In summary, Pst resistance gene Yr78 provides wheat breeders with a new tool to control the devastating stripe rust disease. Since Yr78 is in the same chromosome arm as are Yr35 and Yr36 it would be useful to combine Yr78 with Yr35 and Yr36 in phase to facilitate their simultaneous introgression into commercial wheat varieties.
Author contribution statement
ZD evaluated the populations, analyzed the data and wrote the first draft. JH performed the crosses, developed the mapping populations and contributed to the QTL analysis. JZ contributed to the statistical and QTL analyses. WZ contributed to the construction of the genetic maps. SC provided the SNP genotyping. XC performed the seedling resistance tests and the determination of the Pst races present in the field. YZ supervised ZD and revised the manuscript. JD suggested the project, contributed to the genetic map and statistical analyses and generated the final version of the manuscript. All authors revised the manuscript and provided suggestions.
References
Ali S, Gladieux P, Leconte M, Gautier A, Justesen AF, Hovmøller MS, Enjalbert J, de Vallavieille-Pope C (2014) Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f. sp. tritici. PLoS Pathog 10:e1003903
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic-linkage maps. Genome 36:181–186
Bariana HS, Bansal UK, Schmidt A, Lehmensiek A, Kaur J, Miah H, Howes N, McIntyre CL (2010) Molecular mapping of adult plant stripe rust resistance in wheat and identification of pyramided QTL genotypes. Euphytica 176:251–260
Bulli P, Zhang J, Chao S, Chen X, Pumphrey M (2016) Genetic architecture of resistance to stripe rust in a global winter wheat germplasm collection. G3 (Bethesda) 6:2237–2253
Cavanagh CR, Chao SM, Wang SC, Huang BE, Stephen S, Kiani S, Forrest K, Saintenac C, Brown-Guedira GL, Akhunova A, See D, Bai GH, Pumphrey M, Tomar L, Wong DB, Kong S, Reynolds M, da Silva ML, Bockelman H, Talbert L, Anderson JA, Dreisigacker S, Baenziger S, Carter A, Korzun V, Morrell PL, Dubcovsky J, Morell MK, Sorrells ME, Hayden MJ, Akhunov E (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci USA 110:8057–8062
Chao SM, Dubcovsky J, Dvorak J, Luo MC, Baenziger SP, Matnyazov R, Clark DR, Talbert LE, Anderson JA, Dreisigacker S, Glover K, Chen JL, Campbell K, Bruckner PL, Rudd JC, Haley S, Carver BF, Perry S, Sorrells ME, Akhunov ED (2010) Population- and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.). BMC Genom 11:727
Chen XM (2005) Epidemiology and control of stripe rust (Puccinia striiformis f. sp tritici) on wheat. Can J Plant Pathol 27:314–337
Chen XM (2013) High-temperature adult-plant resistance, key for sustainable control of stripe rust. Am J Plant Sci 4:608–627
Chen XM, Moore M, Milus EA, Long DL, Line RF, Marshall D, Jackson L (2002) Wheat stripe rust epidemics and races of Puccinia striiformis f. sp. tritici in the United States in 2000. Plant Dis 86:39–46
Chen XM, Soria MA, Yan G, Sun J, Dubcovsky J (2003) Development of user-friendly PCR markers for wheat stripe rust resistance gene Yr5. Crop Sci 43:2058–2064
Dadkhodaie NA, Karaoglou H, Wellings CR, Park RF (2011) Mapping genes Lr53 and Yr35 on the short arm of chromosome 6B of common wheat with microsatellite markers and studies of their association with Lr36. Theor Appl Genet 122:479–487
Dedryver F, Paillard S, Mallard S, Robert O, Trottet M, Negre S, Verplancke G, Jahier J (2009) Characterization of genetic components involved in durable resistance to stripe rust in the bread wheat ‘Renan’. Phytopathology 99:968–973
Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J (2009) A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323:1357–1360
Gou JY, Li K, Wu KT, Wang XD, Lin HQ, Cantu D, Uauy C, Dobon-Alonso A, Midorikawa T, Inoue K, Sanchez J, Fu DL, Blechl A, Wallington E, Fahima T, Meeta M, Epstein L, Dubcovsky J (2015) Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell 27:1755–1770
Hovmøller MS, Yahyaoui AH, Milus EA, Justesen AF (2008) Aggressive strains of the wheat yellow rust fungus spread world-wide. Phytopathology 98:S69–S69
Hovmøller MS, Walter S, Bayles RA, Hubbard A, Flath K, Sommerfeldt N, Leconte M, Czembor P, Rodriguez-Algaba J, Thach T, Hansen JG, Lassen P, Justesen AF, Ali S, de Vallavieille-Pope C (2016) Replacement of the European wheat yellow rust population by new races from the centre of diversity in the near-Himalayan region. Plant Pathol 65:402–411
Kolmer JA, Lagudah ES, Lillemo M, Lin M, Bai G (2015) The Lr46 gene conditions partial adult-plant resistance to stripe rust, stem rust, and powdery mildew in Thatcher wheat. Crop Sci 55:2557–2565
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, HuertaEspino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363
Lan CX, Liang SS, Zhou XC, Zhou G, Lu QL, Xia XC, He ZH (2010) Identification of genomic regions controlling adult-plant stripe rust resistance in Chinese landrace Pingyuan 50 through bulked segregant analysis. Phytopathology 100:313–318
Lan CX, Rosewarne GM, Singh RP, Herrera-Foessel SA, Huerta-Espino J, Basnet BR, Zhang YL, Yang EN (2014) QTL characterization of resistance to leaf rust and stripe rust in the spring wheat line Francolin#1. Mol Breed 34:789–803
Lincoln S, Daly M, Lander E (1992) Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical Report 3rd Edition
Line RF, Qayoum A (1992) Virulence, aggressiveness, evolution and distribution of races of Puccinia striiformis (the cause of stripe rust of wheat) in North America, 1968–1987. Technical Volume 1788. United States Department of Agriculture, Washington, p 44
Lowe I, Cantu D, Dubcovsky J (2011) Durable resistance to the wheat rusts: integrating systems biology and traditional phenotype-based research methods to guide the deployment of resistance genes. Euphytica 179:69–79
Maccaferri M, Zhang J, Bulli P, Abate Z, Chao S, Cantu D, Bossolini E, Chen X, Pumphrey M, Dubcovsky J (2015) A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3 (Bethesda) 5:449–465
Marais GF, Pretorius ZA, Wellings CR, McCallum B, Marais AS (2005) Leaf rust and stripe rust resistance genes transferred to common wheat from Triticum dicoccoides. Euphytica 143:115–123
Markell SG, Milus EA (2008) Emergence of a novel population of Puccinia striiformis f. sp. tritici in Eastern United States. Phytopathology 98:632–639
McIntosh RA, Wellings CR, Park RF (1995) Wheat rusts, an atlas of resistance genes. CSIRO, Melbourne
Milus EA, Kristensen K, Hovmøller MS (2008) Increased aggressiveness of Puccinia striiformis f. sp tritici at least partially explains recent stripe rust epidemics. Phytopathology 98:S107–S107
Naruoka Y, Garland-Campbell KA, Carter AH (2015) Genome-wide association mapping for stripe rust (Puccinia striiformis f. sp. tritici) in US Pacific Northwest winter wheat (Triticum aestivum L.). Theor Appl Genet 128:1083–1101
Pahalawatta V, Chen XM (2005) Inheritance and molecular mapping of barley genes conferring resistance to wheat stripe rust. Phytopathology 95:884–889
Riley R, Coucoli H, Chapman V (1967) Chromosomal interchanges and the phylogeny of wheat. Heredity 22:233–247
Santra DK, Chen XM, Santra M, Campbell KG, Kidwell KK (2008) Identification and mapping QTL for high-temperature adult-plant resistance to stripe rust in winter wheat (Triticum aestivum L.) cultivar ‘Stephens’. Theor Appl Genet 117:793–802
Sears ER (1953) Nullisomic analysis in common wheat. Am Nat 87:245–252
Semagn K, Babu R, Hearne S, Olsen M (2014) Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed 33:1–14
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Van Ooijen J (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen 33:10–1371
Wan AM, Chen XM (2014) Virulence characterization of Puccinia striiformis f. sp. tritici using a new set of Yr single-gene line differentials in the United States in 2010. Plant Dis 98:1534–1542
Wang S, Basten CJ, Zeng ZB (2012) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh
Wellings CR (2011) Global status of stripe rust: a review of historical and current threats. Euphytica 179:129–141
William HM, Singh RP, Huerta-Espino J, Palacios G, Suenaga K (2006) Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 49:977–990
Yaniv E, Raats D, Ronin Y, Korol AB, Grama A, Bariana H, Dubcovsky J, Schulman AH, Fahima T (2014) Evaluation of marker-assisted selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Mol Breed 35:43
Acknowledgements
This project was supported by USDA-NRI Grants 2011-68002-30029 (TCAP) and 2017-67007-25939 (WheatCAP) from the USDA National Institute of Food and Agriculture (NIFA) and by the Howard Hughes Medical Institute. ZD was supported by China Scholarship Council Award Number 201406910029.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Statement of human and animal rights
This study did not include human or animal subjects.
Additional information
Communicated by Evans Lagudah.
Electronic supplementary material
Below is the link to the electronic supplementary material.
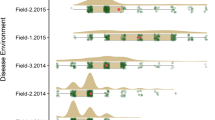
Supplementary Fig. S1. a-j Adult plant resistance in the field for lines validated for Yr78 (QYr.ucw-6B) compared with susceptible control Avocet ‘S’. “F1″ indicate hybrids between the PI accession carrying Yr78 and Avocet ‘S’. k Seedling resistance test in a growth chamber inoculated with field spores. The first three leaves are from plants carrying the all-stage resistance gene Yr35, the fourth leaf is from the susceptible control Avocet ‘S’, and the four last leaves are from BC1F2 plants from population PI 519805/2*Avocet ‘S’ homozygous for Yr78 (and homozygous for the QYr.ucw-1B susceptibility allele).
Supplementary Fig. S2. Linkage map of population PI 519805 × Avocet ‘S’. The 5B and 7B maps were linked due to a reciprocal translocation and were manually separated. For groups of completely linked SNP markers, only one member of the group is presented for simplicity.
Supplementary Fig. S3. QTL analysis of chromosome 1BL in population PI 519805 × Avocet ‘S’ and its comparison with the QTL map of Yr29 in RILs from the population Tc*3/CI13227 (Kolmer et al. 2015).
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Dong, Z., Hegarty, J.M., Zhang, J. et al. Validation and characterization of a QTL for adult plant resistance to stripe rust on wheat chromosome arm 6BS (Yr78). Theor Appl Genet 130, 2127–2137 (2017). https://doi.org/10.1007/s00122-017-2946-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2946-9