Abstract
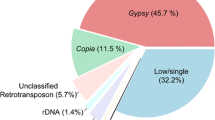
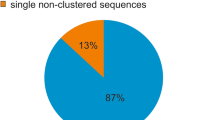
A sample-sequencing strategy combined with slot–blot hybridization and FISH was used to study the composition of the repetitive component of the sunflower genome. One thousand six hundred thirty-eight sequences for a total of 954,517 bp were analyzed. The fraction of sequences that can be classified as repetitive using computational and hybridization approaches amounts to 62% in total. Almost two thirds remain as yet uncharacterized in nature. Of those characterized, most belong to the gypsy superfamily of LTR-retrotransposons. Unlike in other species, where single families can account for large fractions of the genome, it appears that no transposon family has been amplified to very high levels in sunflower. All other known classes of transposable elements were also found. One family of unknown nature (contig 61) was the most repeated in the sunflower genome. The evolution of the repetitive component in the Helianthus genus and in other Asteraceae was studied by comparative analysis of the hybridization of total genomic DNAs from these species to the sunflower small-insert library and compared to gene-based phylogeny. Very little similarity is observed between Helianthus species and two related Asteraceae species outside of the genus. Most repetitive elements are similar in annual and perennial Helianthus species indicating that sequence amplification largely predates such divergence. Gypsy-like elements are more represented in the annuals than in the perennials, while copia-like elements are similarly represented, attesting a different amplification history of the two superfamilies of LTR-retrotransposons in the Helianthus genus.









Similar content being viewed by others
References
Brenner S, Elgar G, Sandford R, MacRae A et al (1993) Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature 366:265–268
Cafasso D, Cozzolino S, De Luca P, Chinali G (2003) An unusual satellite DNA from Zamia paucijuga (Cycadales) characterised by two different organisations of the repetitive unit in the plant genome. Gene 311:71–79
Capy P, Gasperi G, Biémont C, Bazin C (2000) Stress and transposable elements: co-evolution or useful parasites? Heredity 85:101–106
Cavallini A, Zolfino C, Cionini G, Cremonini R, Natali L et al (1986) Nuclear DNA changes within Helianthus annuus L.: cytophotometric, karyological and biochemical analyses. Theor Appl Genet 73:20–26
Ceccarelli M, Sarri V, Natali L, Giordani T, Cavallini A et al (2007) Characterization of the chromosome complement of Helianthus annuus by in situ hybridization of a tandemly repeated DNA sequence. Genome 50:429–434
Charles M, Belcram H, Just J, Huneau C, Viollet A et al (2008) Dynamics and differential proliferation of transposable elements during the evolution of the B and A genomes of wheat. Genetics 180:1071–1086
Diaz-Martin J, Almoguera C, Prieto-Dapena P, Espinosa JM, Jordano J (2005) Functional interaction between two transcription factors involved in the developmental regulation of a small heat stress protein gene promoter. Plant Physiol 139:1483–1494
Doyle JJ, Doyle JL (1989) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Elgar G, Clark MS, Meek S, Smith S et al (1999) Generation and analysis of 25 Mb of genomic DNA from the pufferfish Fugu rubripes by sequence scanning. Genome Res 9:960–971
Felsenstein J (1989) PHYLIP: phylogeny inference package. Cladistics 5:164–166
Gindullis F, Desel C, Galasso I, Schmidt T (2001) The large scale organization of the centromeric region in Beta species. Genome Res 11:253–265
Giordani T, Natali L, Cavallini A (2003) Analysis of a dehydrin encoding gene and its phylogenetic utility in Helianthus. Theor Appl Genet 107:316–325
Goff SA, Ricke D, Lan TH, Presting G et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Harter AV, Gardner KA, Falush D, Lentz DL, Bye R et al (2004) Origin of extant domesticated sunflowers in eastern North America. Nature 430:201–205
Hawkins JS, Kim HR, Nason JD, Wing RA, Wendel JF (2006) Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res 16:1252–1261
Heslop-Harrison JS, Brandes A, Taketa S, Schmidt T, Vershinin AV et al (1997) The chromosomal distribution of Ty1-copia group retrotransposable elements in higher plants and their implications for genome evolution. Genetica 100:197–204
Hill P, Burford D, Martin DMA, Flavell AJ (2005) Retrotransposon populations of Vicia species with varying genome size. Mol Genet Genom 273:371–381
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Jiang N, Wessler SR (2001) Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 13:2533–2564
Jiang N, Feschotte C, Zhang X, Wessler SR (2004) Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs). Curr Opin Plant Biol 7:115–119
Lentz DL, Pohl MD, Alvarado JL, Tarighat S, Bye R (2008) Sunflower (Helianthus annuus L.) as a pre-Columbian domesticate in Mexico. Proc Natl Acad Sci USA 105:6232–6237
Li W, Zhang P, Fellers JP, Friebe B, Gill BS (2004) Sequence composition, organization, and evolution of the core Triticeae genome. Plant J 40:500–511
Liu Z, Yue W, Li D, Wang RR, Kong X, Lu K, Wang G, Dong Y, Jin W, Zhang X (2008) Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma 117:445–456
Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 34:12404–12410
Macas J, Neumann P, Navrátilová A (2007) Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 8:427–442
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Meyers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11:1660–1676
Neumann P, Koblizkova A, Navratilova A, Macas J (2006) Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 173:1047–1056
Ouvrard O, Cellier F, Ferrare K, Tousch D, Lamaze T et al (1996) Identification and expression of water stress- and abscisic acid-regulated genes in a drought-tolerant sunflower genotype. Plant Mol Biol 31:819–829
Paux E, Roger D, Badaeva E, Gay G, Bernard M et al (2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J 48:463–474
Pearce SR, Harrison G, Li D, Heslop-Harrison J et al (1996) The Ty1-copia group retrotransposons in Vicia species: Copy number, sequence heterogeneity and chromosomal localisation. Mol Gen Genet 250:305–315
Pich U, Schubert I (1998) Terminal heterochromatin and alternative telomeric sequences in Allium cepa. Chromosome Res 6:315–321
Piegu B, Guyot R, Picault N, Roulin A, Saniyal A et al (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269
Presting GG, Malysheva L, Fuchs J, Schubert I (1998) A Ty3/gypsy retrotransposon-like sequence localized to the centromeric regions of cereal chromosomes. Plant J 16:721–728
Rabinowicz D, Citek R, Budiman MA, Nunberg A, Bedell JA et al (2005) Differential methylation of genes and repeats in land plants. Genome Res 15:1431–1440
Rieseberg LH (1995) The role of hybridization in evolution: old wine in new skins. Am J Bot 82:944–953
Rieseberg LH, Beckstrom-Sternberg SM, Liston A, Arias DM (1991) Phylogenetic and systematic inferences from chloroplast DNA and isozyme variation in Helianthus sect. Helianthus (Asteraceae). Syst Bot 16:50–76
Rohlf FJ (1998) NTSYS-pc. Numerical taxonomy and multivariate analysis system (version 2.02 j). Exeter Software, Setauket
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
SanMiguel P, Tikhonov A, Springer PS, Edwards KJ, Lee M et al (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768
Santini S, Cavallini A, Natali L, Minelli S et al (2002) Ty1/copia- and Ty3/gypsy-like DNA sequences in Helianthus species. Chromosoma 111:192–200
Schilling EE (1997) Phylogenetic analysis of Helianthus (Asteraceae) based on chloroplast DNA restriction-site data. Theor Appl Genet 94:925–933
Schilling EE (2001) Phylogeny of Helianthus and related genera. Oleagineaux Corps Gras Lipides 8:22–25
Schilling EE, Heiser CB (1981) Infrageneric classification of Helianthus (Compositae). Taxonomy 30:393–403
Schilling EE, Linder CR, Noyes RD, Rieseberg LH (1998) Phylogenetic relationships in Helianthus (Asteraceae) based on nuclear ribosomal DNA internal transcribed spacer region sequence data. Syst Bot 23:177–187
Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS (1989) In situ localization of parental genomes in a wide hybrid. Ann Bot 64:315–324
Soltis ED, Soltis PS (2000) Contributions of plant molecular systematics to studies of molecular evolution. Plant Mol Biol 42:45–75
Sossey-Alaoui K, Serieys H, Tersac M, Lambert P, Schilling EE et al (1998) Evidence for several genomes in Helianthus. Theor Appl Genet 97:422–430
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
The French-Italian Public Consortium for grape genome characterization (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
The International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
The International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Thompson JD, Desmond G, Gibson H, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Timme RE, Simpson BB, Linder CR (2007) High-resolution phylogeny for Helianthus (Asteraceae) using the 18 s–26 s ribosomal DNA external transcribed spacer. Am J Bot 94:1837–1852
Tuskan GA, DiFazio S, Jansson S, Bohlmann J et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Vicient CM, Kalendar R, Anamthawat-Jónsson K, Suoniemi A, Schulman AH (1999) Structure, functionality, and evolution of the BARE-1 retrotransposon of barley. Genetica 107:53–63
Vicient CM, Kalendar R, Schulman AH (2005) Variability, recombination, and mosaic evolution of the barley BARE-1 retrotransposon. J Mol Evol 61:275–291
Vitte C, Bennetzen JL (2006) Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Nat Acad Sci USA 103:17638–17643
Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B (2001) Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J 26:307–316
Wicker T, Sabot F, Hua-Van A, Bennetzen JL et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Wilson RK, Mardis ER (1997) Genome analysis: a laboratory manual. vol 1. Analyzing DNA. Cold Spring Harbor Laboratory Press, New York
Zhang D, Yang Q, Bao W, Zhang Y, Han B et al (2005) Molecular cytogenetic characterization of the Antirrhinum majus genome. Genetics 169:325–335
Acknowledgments
This work was supported by PRIN-MIUR, Projects “Caratterizzazione della componente ripetitiva di genomi complessi in specie vegetali: modelli per angiosperme e gimnosperme” and “Variabilità di sequenza ed eterosi in piante coltivate”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Bervillé.
Sequences from Helianthus annuus randomly sheared genomic DNA library and sequences of genes used for phylogenetic analyses are available at the URL: https://services.appliedgenomics.org/sequences-export/26-Helianthus/.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cavallini, A., Natali, L., Zuccolo, A. et al. Analysis of transposons and repeat composition of the sunflower (Helianthus annuus L.) genome. Theor Appl Genet 120, 491–508 (2010). https://doi.org/10.1007/s00122-009-1170-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1170-7