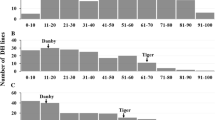
Abstract
The premature germination of seeds before harvest, known as preharvest sprouting (PHS), is a serious problem in all wheat growing regions of the world. In order to determine genetic control of PHS resistance in white wheat from the relatively uncharacterized North American germplasm, a doubled haploid population consisting of 209 lines from a cross between the PHS resistant variety Cayuga and the PHS susceptible variety Caledonia was used for QTL mapping. A total of 16 environments were used to detect 15 different PHS QTL including a major QTL, QPhs.cnl-2B.1, that was significant in all environments tested and explained from 5 to 31% of the trait variation in a given environment. Three other QTL QPhs.cnl-2D.1, QPhs.cnl-3D.1, and QPhs.cnl-6D.1 were detected in six, four, and ten environments, respectively. The potentially related traits of heading date (HD), plant height (HT), seed dormancy (DOR), and rate of germination (ROG) were also recorded in a limited number of environments. HD was found to be significantly negatively correlated with PHS score in most environments, likely due to a major HD QTL, QHd.cnl-2B.1, found to be tightly linked to the PHS QTL QPhs.cnl-2B.1. Using greenhouse grown material no overlap was found between seed dormancy and the four most consistent PHS QTL, suggesting that greenhouse environments are not representative of field environments. This study provides valuable information for marker-assisted breeding for PHS resistance, future haplotyping studies, and research into seed dormancy.

Similar content being viewed by others
References
Anderson JA, Ogihara Y, Sorrells ME, Tanksley SD (1992) Development of a chromosomal arm map for wheat based on RFLP markers. Theor Appl Genet 83:1035–1043
Anderson JA, Sorrells ME, Tanksley SD (1993) RFLP analysis of genomic regions associated with resistance to preharvest sprouting in wheat. Crop Sci 33:453–459
Bailey PC, Mckibbin RS, Lenton JR, Holdsworth MJ, Flintham JE, Gale MD (1999) Genetic map locations for orthologous Vp1 genes in wheat and rice. Theor Appl Genet 98:281–284
Basten CJ, Weir BS, Zeng ZB (1994) Zmap—a QTL Cartographer. In: Smith C, Gavora JS, Benkel B, Chesnais J, Fairfull W, Gibson JP et al (eds) Proceedings of the 5th World congress on genetics applied to livestock production: Computing Strategies and Software, Guelph, Ontario, Canada
Basten CJ, Weir BS, Zeng ZB (2002) QTL Cartographer. Department of Statistics, North Carolina State University, Raleigh, NC
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115:721–733
Bewley JD, Black M (1994) Seeds:Physiology of Development and Germination. Plenum Press, New York
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Ellis H, Spielmeyer W, Gale R, Rebetzke J, Richards A (2002) “Perfect” markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet 105:1038–1042
Flintham J (2000) Different genetic components control coat-imposed and embryo-imposed dormancy in wheat. Seed Sci Res 10:43–50
Flintham J, Adlam R, Bassoi M, Holdsworth M, Gale M (2002) Mapping genes for resistance to sprouting damage in wheat. Euphytica 126:39–45
Groos C, Gay G, Perretant M, Gervais L, Bernard M, Dedryver F et al (2002) Study of the relationship between pre-harvest sprouting and grain color by quantitative trait loci analysis in a white × red grain bread-wheat cross. Theor Appl Genet 104:39–47
Gupta PK, Balyan HS, Edwards KJ, Isaac P, Korzun V, Röder M et al (2002) Genetic mapping of 66 new microstalleite (SSR) loci in bread wheat. Theor Appl Genet 105:413–422
Guyomarc’h H, Charmet G, Edwards KJ, Bernard M (2002) Characterisation of polymorphic microsatellite markers from Aegilops tauschii and transferability to the D-genome of bread wheat. Theor Appl Genet 104:1164–1172
Hanocq E, Niarquin M, Heumez E, Rousset M, Le Gouis J (2004) Detection and mapping of QTL for earliness components in a bread wheat recombinant inbred lines population. Theor Appl Genet 110:106–115
Heun M, Kennedy AE, Anderson JA, Lapitan NLV, Sorrells ME, Tanksley SD (1991) Construction of a restriction fragment length polymorphism map for barley (Hordeum vulgare). Genome 34:437–447
Imtiaz M, Ogbonnaya FC, Oman J, Van Ginkel M (2008) Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines. Genetics 178:1725–1736
Kato K, Nakamura W, Tabiki T, Miura H (2001) Detection of loci controlling seed dormancy on group 4 chromosomes of wheat and comparative mapping with rice and barley genomes. Theor Appl Genet 10:980–985
King RW, Richards RA (1984) Water uptake in relation to pre-harvest sprouting damage in wheat:Ear characteristics. Aust J Agric Res 35:327–336
King RW, Von Wettstein-Knowles P (2000) Epicuticular waxes and regulation of ear wetting and pre-harvest sprouting in barley and wheat. Euphytica 112:157–166
Kulwal PL, Singh R, Balyan HS, Gupta PK (2004) Genetic basis of pre-harvest sprouting tolerance using single-locus and two-locus QTL analyses in bread wheat. Funct Integr Genomics 4:94–101
Kulwal PL, Kumar N, Gaur A, Khurana P, Khurana JP, Tyagi AK et al (2005) Mapping of a major QTL for pre-harvest sprouting tolerance on chromosome 3A in bread wheat. Theor Appl Genet 111:1052–1059
Liu ZH, Anderson JA, Hu J, Friesen TL, Rasmussen JB, Faris JD (2005) A wheat intervarietal genetic linkage map based on microsatellite and target region amplified polymorphism markers and its utility for detecting quantitative trait loci. Theor Appl Genet 111:782–794
Liu S, Cai S, Graybosch R, Chen C, Bai G (2008) Quantitative trait loci for resistance to pre-harvest sprouting in US hard white winter wheat Rio Blanco. Theor Appl Genet 117:691–699
Mangin B, Goffinet B, Rebai A (1994) Constructing confidence intervals for QTL location. Genetics 138:1301–1308
Manly KF, Cudmore RH, Meer M (2001) Map manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932
Mares DJ, Mrva K (2001) Mapping quantitative trait loci associated with variation in grain dormancy in Australian wheat. Aust J Agric Res 52:1257–1265
Mares DJ, Mrva K, Tan MK, Sharp P (2002) Dormancy in white-grained wheat:Progress towards identification of genes and molecular markers. Euphytica 126:47–53
Mares D, Mrva K, Cheong J, Williams K, Watson B, Storlie E et al (2005) A QTL located on chromosome 4A associated with dormancy in white- and red-grained wheats of diverse origin. Theor Appl Genet 111:1357–1364
Miura H, Sato N, Kato K, Amano Y (2002) Detection of chromosomes carrying genes for seed dormancy of wheat using the backcross reciprocal monosomic method. Plant Breeding 121:394–399
Mori M, Uchino N, Chono M, Kato K, Miura H (2005) Mapping QTLs for grain dormancy on wheat chromosome 3A and the group 4 chromosomes, and their combined effect. Theor Appl Genet 110:1257–1266
Mrva K, Mares DJ (1996) Expression of late maturity alpha-amylase in wheat containing gibberellic acid insensitivity genes. Euphytica 88:69–76
Ogbonnaya FC, Imtiaz M, Depauw RM (2007) Haplotype diversity of preharvest sprouting QTLs in wheat. Genome 50:107–118
Ogbonnaya FC, Imtiaz M, Ye G, Hearnden PR, Hernandez E, Eastwood RF et al (2008) Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor Appl Genet 116:891–902
Osa M, Kato K, Mori M, Shindo C, Torada A, Miura H (2003) Mapping QTLs for seed dormancy and the Vp1 homologue on chromosome 3A in wheat. Theor Appl Genet 106:1491–1496
Pestsova E, Ganal MW, Röder MS (2000) Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 43:689–697
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier M, Leroy P et al (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Roy J, Prasad M, Varshney R, Balyan H, Blake T, Dhaliwal HS et al (1999) Identification of a microsatellite on chromosomes 6B and a STS on 7D of bread wheat showing an association with preharvest sprouting tolerance. Theor Appl Genet 99:336–340
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J et al (2005) Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550–560
Sourdille P, Gandon B, Chiquet V, Nicot N, Somers D, Murigneux A et al (2004) Wheat Genoplante SSR mapping data release:A new set of markers and comprehensive genetic and physical mapping data. Graingenes. http://wheat.pw.usda.gov
Tan MK, Sharp PJ, Lu MQ, Howes N (2006) Genetics of grain dormancy in a white wheat. Aust J Agr Res 57:1157–1165
Torada A, Amano Y (2002) Effect of seed coat color on seed dormancy in different environments. Euphytica 126:99–105
Torada A, Koike M, Ikeguchi S, Tsutsui I (2008) Mapping of a major locus controlling seed dormancy using backcrossed progenies in wheat (Triticum aestivum L.). Genome 51:426–432
Vos P, Hogers R, Bleeker M, Reijans M, Van De Lee T et al (1995) AFLP:A new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Walker-Simmons M (1987) ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol 84:61–66
Walker-Simmons M (1988) Enhancement of ABA responsiveness in wheat embryos by high temperature. Plant Cell Environ 11:769–775
Wu J, Carver BF, Goad CL (1999) Kernel color variability of hard white and hard red winter wheat. Crop Sci 39:634–638
Yang J, Zhu J, Williams RW (2007a) Mapping the genetic architecture of complex traits in experimental populations. Bioinformatics 23:1527–1536
Yang Y, Zhao XL, Xia LQ, Chen XM, Xia XC, Yu Z et al (2007b) Development and validation of a Viviparous-1 STS marker for pre-harvest sprouting tolerance in Chinese wheats. Theor Appl Genet 115:971–980
Yang J, Hu C, Hu H, Yu R, Xia Z, Ye X et al (2008) QTLNetwork:Mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24:721–723
Yu J, Dake TM, Singh S, Benscher D, Li W, Gill B et al (2004) Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome 47:805–818
Zanetti S, Winzeler M, Keller M, Keller B, Messmer M (2000) Genetic analysis of pre-harvest sprouting resistance in a wheat × spelt cross. Crop Sci 40:1406–1417
Acknowledgments
This research was supported by National Research Initiative Competitive Grants 2005-35301-15728 and 2006-55606-16629 from the USDA Cooperative State Research, Education, and Extension Service and Hatch project, pp 149–419.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Romagosa.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Munkvold, J.D., Tanaka, J., Benscher, D. et al. Mapping quantitative trait loci for preharvest sprouting resistance in white wheat. Theor Appl Genet 119, 1223–1235 (2009). https://doi.org/10.1007/s00122-009-1123-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1123-1