Abstract
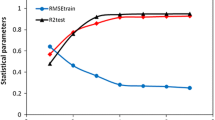
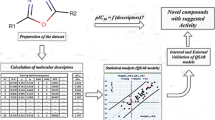
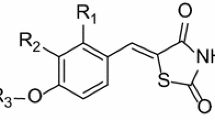
In this study, the quantitative structure–activity relationship (QSAR) model for some pyrazole/imidazole amide derivatives as mGlu5 inhibitors was developed. The data set was split into the training and test subsets, randomly. The most relevant variables were selected using the genetic algorithm (GA) variable selection method. Multiple linear regression (MLR) method was used as a linear model to predict the activity of mGlu5 inhibitors based on compounds in training set. The external set of nine compounds selected out of 47 compounds, and used to evaluate the predictive ability of QSAR model. The built model could give high statistical quantities (R 2train = 0.837, Q 2 = 0.759, R 2test = 0.919) in which proved that the GA-MLR model was a useful tool to predict the inhibitory activity of pyrazole/imidazole amide derivatives. The results suggested that the atomic masses, atomic van der Waals volumes, atomic electronegativities, and the number of imines (aromatic) are the most important independent factors that contribute to the mGlu5 inhibition activity of pyrazole/imidazole amides derivatives.


Similar content being viewed by others
References
Adimi M, Salimi M, Nekoei M, Pourbasheer E, Beheshti A (2012) A quantitative structure-activity relationship study on histamine receptor antagonists using the genetic algorithm-multi-parameter linear regression method. J Serb Chem Soc 77:639–650
Afantitis A, Melagraki G, Sarimveis H, Koutentis P, Markopoulos J, Igglessi-Markopoulou O (2006) A novel simple QSAR model for the prediction of anti-HIV activity using multiple linear regression analysis. Mol Divers 10:405–414
Agrawal VK, Khadikar PV (2001) QSAR prediction of toxicity of nitrobenzenes. Bioorg Med Chem 9:3035–3040
Beheshti A, Pourbasheer E, Nekoei M, Vahdani S (2012) QSAR modeling of antimalarial activity of urea derivatives using genetic algorithm-multiple linear regressions. J Saudi Chem Soc. doi:10.1016/j.jscs.2012.07.019
Bhave G, Karim F, Carlton SM, Gereau Iv RW (2001) Peripheral group I metabotropic glutamate receptors modulate nociception in mice. Nat Neurosci 4:417–423
Bonnefous C, Vernier J-M, Hutchinson JH, Chung J, Reyes-Manalo G, Kamenecka T (2005) Dipyridyl amides: potent metabotropic glutamate subtype 5 (mGlu5) receptor antagonists. Bioorg Med Chem Lett 15:1197–1200
Burbidge R, Trotter M, Buxton B, Holden S (2001) Drug design by machine learning: support vector machines for pharmaceutical data analysis. Comput Chem 26:5–14
Chae E, Shin Y-J, Ryu E-J, Ji MK, Ryune Cho N, Lee K-H, Jeong HJ, Kim S-J, Choi Y, Seok OhK, Park C-E, Soo Yoon Y (2013) Discovery of biological evaluation of pyrazole/imidazole amides as mGlu5 receptor negative allosteric modulators. Bioorg Med Chem Lett 23:2134–2139
Chaki S, Ago Y, Palucha-Paniewiera A, Matrisciano F, Pilc A (2013) mGlu2/3 and mGlu5 receptors: potential targets for novel antidepressants. Neuropharmacology 66:40–52
Conn PJ, Pin J-P (1997) Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol 37:205–237
Emmitte KA (2011) Recent advances in the design and development of novel negative allosteric modulators of mGlu5. ACS Chem Neurosci 2:411–432
Eriksson L, Johansson E, Müller M, Wold S (2000) On the selection of the training set in environmental QSAR analysis when compounds are clustered. J Chemom 14:599–616
Firoozpour L, Sadatnezhad K, Dehghani S, Pourbasheer E, Foroumadi A, Shafiee A, Amanlou M (2012) An efficient piecewise linear model for predicting activity of caspase-3 inhibitors. DARU J Pharm Sci. doi:10.1186/2008-2231-20-31
Ghasemi J, Saaidpour S (2007) Quantitative structure–property relationship study of n-octanol–water partition coefficients of some of diverse drugs using multiple linear regression. Anal Chim Acta 604:99–106
Habibi-Yangjeh A, Pourbasheer E, Danandeh-Jenagharad M (2008a) Prediction of basicity constants of various pyridines in aqueous solution using a principal component-genetic algorithm-artificial neural network. Mon Chem 139:1423–1431
Habibi-Yangjeh A, Pourbasheer E, Danandeh-Jenagharad M (2008b) Prediction of melting point for drug-like compounds using principal component-genetic algorithm-artificial neural network. Bull Korean Chem Soc 29:833–841
Habibi-Yangjeh A, Pourbasheer E, Danandeh-Jenagharad M (2009) Application of principal component-genetic algorithm-artificial neural network for prediction acidity constant of various nitrogen-containing compounds in water. Mon Chem 140:15–27
Holland JH (1975) Adaptation in natural and artificial systems. University of Michigan Press, Ann Arbor
HyperChem (2002) Molecular modeling system, 7.03rd edn. Hypercube, Gainesville
Jeffrey Conn P, Christopoulos A, Lindsley CW (2009) Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov 8:41–54
Leardi R, Boggia R, Terrile M (1992) Genetic algorithms as a strategy for feature selection. J Chemom 6:267–281
Lindsley CW, Bates BS, Menon UN, Jadhav SB, Kane AS, Jones CK, Rodriguez AL, Conn PJ, Olsen CM, Winder DG, Emmitte KA (2011) (3-Cyano-5-fluorophenyl)biaryl negative allosteric modulators of mGlu5: discovery of a new tool compound with activity in the OSS mouse model of addiction. ACS Chem Neurosci 2:471–482
Mathworks (2005) Genetic algorithm and direct search toolbox users guide. The Mathworks
Pin JP, Duvoisin R (1995) The metabotropic glutamate receptors: structure and functions. Neuropharmacology 34:1–26
Pourbasheer E, Aalizadeh R, Ganjali, Norouzi P (2013a) QSAR study of IKKβ inhibitors by the genetic algorithm: multiple linear regressions. Med Chem Res. doi:10.1007/s00044-013-0611-7
Pourbasheer E, Aalizadeh R, Ganjali, Norouzi P (2013b) QSAR study of α1β4 integrin inhibitors by GA-MLR and GA-SVM methods. Struct Chem. doi:10.1007/s11224-013-0300-7
Pourbasheer E, Aalizadeh R, Ganjali MR, Norouzi P, Shadmanesh J, Methenitis C (2013c) QSAR study of Nav1.7 antagonists by multiple linear regression method based on genetic algorithm (GA-MLR). Med Chem Res. doi:10.1007/s00044-013-0821-z
Pourbasheer E, Ahmadpour S, Zare-Dorabei R, Nekoei M (2013d) Quantitative structure activity relationship study of p38α MAP kinase inhibitors. Arab J Chem. doi:10.1016/j.arabjc.2013.05.009
Pourbasheer E, Beheshti A, Khajehsharifi H, Ganjali MR, Norouzi P (2013e) QSAR study on hERG inhibitory effect of kappa opioid receptor antagonists by linear and non-linear methods. Med Chem Res 22:4047–4058
Saugstad J, Ingram S (2008) Group I metabotropic glutamate receptors (mGlu1 and mGlu5). In: Gereau R, Swanson G (eds) The glutamate receptors. The receptors. Humana, Totawa, pp 387–463
Sevostianova N, Danysz W (2006) Analgesic effects of mGlu1 and mGlu5 receptor antagonists in the rat formalin test. Neuropharmacology 51:623–630
Storto M, Ngomba RT, Battaglia G, Freitas I, Griffini P, Richelmi P, Nicoletti F, Vairetti M (2003) Selective blockade of mGlu5 metabotropic glutamate receptors is protective against acetaminophen hepatotoxicity in mice. J Hepatol 38:179–187
Timmerman H (1995) New developments and applications: QSAR and drug design. In: Fujita T (ed) Pharmacochemistry library. Elsevier, Amsterdam, pp 413–450
Todeschini R, Consonni V (2008) Handbook of molecular descriptors. Wiley, Weinheim
Todeschini R, Consonni V, Mauri A, Pavan M (2005) DRAGON software for the calculation of molecular descriptors, 5.3rd edn. Talete SRL, Milan
Tropsha A, Gramatica P, Gombar VK (2003) The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Comb Sci 22:69–77
Walker K, Reeve A, Bowes M, Winter J, Wotherspoon G, Davis A, Schmid P, Gasparini F, Kuhn R, Urban L (2001) mGlu5 receptors and nociceptive function II. mGlu5 receptors functionally expressed on peripheral sensory neurones mediate inflammatory hyperalgesia. Neuropharmacology 40:10–19
Waller CL, Bradley MP (1999) Development and validation of a novel variable selection technique with application to multidimensional quantitative structure–activity relationship studies. J Chem Inf Comput Sci 39:345–355
Zhou S, Komak S, Du J, Carlton SM (2001) Metabotropic glutamate 1α receptors on peripheral primary afferent fibers: their role in nociception. Brain Res 913:18–26
Acknowledgments
The authors would like to thank Young Researchers and Elite Club and the State Scholarships’ Foundation of Greece (I.K.Y.) for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pourbasheer, E., Aalizadeh, R., Ganjali, M.R. et al. QSAR study of mGlu5 inhibitors by genetic algorithm-multiple linear regressions. Med Chem Res 23, 3082–3091 (2014). https://doi.org/10.1007/s00044-013-0896-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-013-0896-6