Abstract
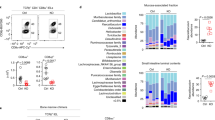
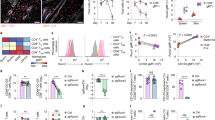
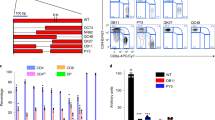
Integrin CD103 mediates the adhesion and tissue retention of T cells by binding to E-cadherin which is abundant on epithelial cells. Notably, CD103 is highly expressed on CD8 T cells but conspicuously absent on most CD4 T cells. The mechanism controlling such lineage-specific expression of CD103 remains unclear. Using a series of genetically engineered mouse models, here, we demonstrate that the regulatory mechanism of CD103 expression is distinct between CD4 and CD8 T cells, and that the transcription factor Runx3 plays an important but not an essential role in this process. We further found that the availability of integrin β7 which heterodimerizes with CD103 was necessary but also constrained the surface expression of CD103. Notably, the forced surface expression of CD103 did not significantly alter the thymic development of conventional T cells but severely impaired the generation of MHC-II-restricted TCR transgenic T cells, revealing previously unappreciated aspects of CD103 in the selection and maturation of CD4 T cells. Unlike its effect on CD4 T cell development, however, CD103 overexpression did not significantly affect CD4 T cells in peripheral tissues. Moreover, the frequency and number of CD4 T cells in the small intestine epithelium did not increase even though E-cadherin is highly expressed in this tissue. Collectively, these results suggest that most mature CD4 T cells are refractory to the effects of CD103 expression, and that they presumably utilize CD103-independent pathways to control their tissue retention and residency.






Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Singer A, Adoro S, Park JH (2008) Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol 8(10):788–801. https://doi.org/10.1038/nri2416
Taniuchi I (2018) CD4 helper and CD8 cytotoxic T cell differentiation. Annu Rev Immunol 36:579–601. https://doi.org/10.1146/annurev-immunol-042617-053411
Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I (2008) Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319(5864):822–825. https://doi.org/10.1126/science.1151844
Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR (2007) The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med 204(8):1945–1957. https://doi.org/10.1084/jem.20070133
Ehlers M, Laule-Kilian K, Petter M, Aldrian CJ, Grueter B, Wurch A, Yoshida N, Watanabe T, Satake M, Steimle V (2003) Morpholino antisense oligonucleotide-mediated gene knockdown during thymocyte development reveals role for Runx3 transcription factor in CD4 silencing during development of CD4-/CD8+ thymocytes. J Immunol 171(7):3594–3604. https://doi.org/10.4049/jimmunol.171.7.3594
Park J-H, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, Kimura MY, Cui Y, Lucas PJ, Gress RE (2010) Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol 11(3):257
Etzensperger R, Kadakia T, Tai XG, Alag A, Guinter TI, Egawa T, Erman B, Singer A (2017) Identification of lineage-specifying cytokines that signal all CD8(+)-cytotoxic-lineage-fate “decisions” in the thymus. Nat Immunol 18(11):1218–1227. https://doi.org/10.1038/ni.3847
Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, Bernstein Y, Goldenberg D, Brenner O, Berke G, Levanon D, Groner Y (2003) Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A 100(13):7731–7736. https://doi.org/10.1073/pnas.1232420100
Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A (2009) Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med 206(1):51–59. https://doi.org/10.1084/jem.20081242
Shan Q, Zeng Z, Xing S, Li F, Hartwig SM, Gullicksrud JA, Kurup SP, Van Braeckel-Budimir N, Su Y, Martin MD, Varga SM, Taniuchi I, Harty JT, Peng W, Badovinac VP, Xue HH (2017) The transcription factor Runx3 guards cytotoxic CD8(+) effector T cells against deviation towards follicular helper T cell lineage. Nat Immunol 18(8):931–939. https://doi.org/10.1038/ni.3773
Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, Ludwig Y, Fukuyama H, Wardemann H, Waldschuetz R (2005) Runx3 regulates integrin αE/CD103 and CD4 expression during development of CD4−/CD8+ T cells. J Immunol 175(3):1694–1705
Yarmus M, Woolf E, Bernstein Y, Fainaru O, Negreanu V, Levanon D, Groner Y (2006) Groucho/transducin-like Enhancer-of-split (TLE)-dependent and -independent transcriptional regulation by Runx3. Proc Natl Acad Sci U S A 103(19):7384–7389. https://doi.org/10.1073/pnas.0602470103
Egawa T, Littman DR (2008) ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol 9(10):1131–1139. https://doi.org/10.1038/ni.1652
Hermiston ML, Gordon JI (1995) In vivo analysis of cadherin function in the mouse intestinal epithelium: essential roles in adhesion, maintenance of differentiation, and regulation of programmed cell death. J Cell Biol 129(2):489–506. https://doi.org/10.1083/jcb.129.2.489
Schenkel JM, Masopust D (2014) Tissue-resident memory T cells. Immunity 41(6):886–897. https://doi.org/10.1016/j.immuni.2014.12.007
Mowat AM, Agace WW (2014) Regional specialization within the intestinal immune system. Nat Rev Immunol 14(10):667–685. https://doi.org/10.1038/nri3738
Camerini V, Panwala C, Kronenberg M (1993) Regional specialization of the mucosal immune-system - intraepithelial lymphocytes of the large-intestine have a different phenotype and function than those of the small-intestine. J Immunol 151(4):1765–1776
Parrott DM, Tait C, MacKenzie S, Mowat AM, Davies MD, Micklem HS (1983) Analysis of the effector functions of different populations of mucosal lymphocytes. Ann N Y Acad Sci 409(1):307–320. https://doi.org/10.1111/j.1749-6632.1983.tb26879.x
Schön MP, Arya A, Murphy EA, Adams CM, Strauch UG, Agace WW, Marsal J, Donohue JP, Her H, Beier DR (1999) Mucosal T lymphocyte numbers are selectively reduced in integrin αE (CD103)-deficient mice. J Immunol 162(11):6641–6649
Robinson PW, Green SJ, Carter C, Coadwell J, Kilshaw PJ (2001) Studies on transcriptional regulation of the mucosal T-cell integrin alphaEbeta7 (CD103). Immunology 103(2):146–154. https://doi.org/10.1046/j.1365-2567.2001.01232.x
Allakhverdi Z, Fitzpatrick D, Boisvert A, Baba N, Bouguermouh S, Sarfati M, Delespesse G (2006) Expression of CD103 identifies human regulatory T-cell subsets. J Allergy Clin Immunol 118(6):1342–1349. https://doi.org/10.1016/j.jaci.2006.07.034
Bertoni A, Alabiso O, Galetto AS, Baldanzi G (2018) Integrins in T cell physiology. Int J Mol Sci 19(2):485. https://doi.org/10.3390/ijms19020485
Kutlesa S, Wessels JT, Speiser A, Steiert I, Muller CA, Klein G (2002) E-cadherin-mediated interactions of thymic epithelial cells with CD103+ thymocytes lead to enhanced thymocyte cell proliferation. J Cell Sci 115(Pt 23):4505–4515. https://doi.org/10.1242/jcs.00142
Bettelli E, Pagany M, Weiner HL, Linington C, Sobel RA, Kuchroo VK (2003) Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med 197(9):1073–1081. https://doi.org/10.1084/jem.20021603
Yamashita I, Nagata T, Tada T, Nakayama T (1993) Cd69 cell-surface expression identifies developing thymocytes which audition for T-cell antigen receptor-mediated positive selection. Int Immunol 5(9):1139–1150. https://doi.org/10.1093/intimm/5.9.1139
Pabst R, Russell MW, Brandtzaeg P (2008) Tissue distribution of lymphocytes and plasma cells and the role of the gut. Trends Immunol 29(5):206–208. https://doi.org/10.1016/j.it.2008.02.006 (author reply 209–210)
Zhou C, Qiu Y, Yang H (2019) CD4CD8alphaalpha IELs: they have something to say. Front Immunol 10:2269. https://doi.org/10.3389/fimmu.2019.02269
Uehara S, Grinberg A, Farber JM, Love PE (2002) A role for CCR9 in T lymphocyte development and migration. J Immunol 168(6):2811–2819. https://doi.org/10.4049/jimmunol.168.6.2811
Higgins JM, Mandlebrot DA, Shaw SK, Russell GJ, Murphy EA, Chen YT, Nelson WJ, Parker CM, Brenner MB (1998) Direct and regulated interaction of integrin alphaEbeta7 with E-cadherin. J Cell Biol 140(1):197–210. https://doi.org/10.1083/jcb.140.1.197
Smyth L, Kirby J, Cunningham A (2007) Role of the mucosal integrin αE (CD103) β7 in tissue-restricted cytotoxicity. Clin Exp Immunol 149(1):162–170
Fousteri G, Dave A, Juntti T, von Herrath M (2009) CD103 is dispensable for anti-viral immunity and autoimmunity in a mouse model of virally-induced autoimmune diabetes. J Autoimmun 32(1):70–77. https://doi.org/10.1016/j.jaut.2008.12.001
El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, Drachenberg CB, Hadley GA (2005) TGF-{beta}-dependent CD103 expression by CD8(+) T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J Exp Med 201(10):1647–1657. https://doi.org/10.1084/jem.20041044
Feng Y, Wang DH, Yuan RW, Parker CM, Farber DL, Hadley GA (2002) CD103 expression is required for destruction of pancreatic islet allografts by CD8(+) T cells. J Exp Med 196(7):877–886. https://doi.org/10.1084/jem.20020178
Boller K, Vestweber D, Kemler R (1985) Cell-adhesion molecule uvomorulin is localized in the intermediate junctions of adult intestinal epithelial cells. J Cell Biol 100(1):327–332. https://doi.org/10.1083/jcb.100.1.327
Nguyen QP, Deng TZ, Witherden DA, Goldrath AW (2019) Origins of CD4(+) circulating and tissue-resident memory T-cells. Immunology 157(1):3–12. https://doi.org/10.1111/imm.13059
Wurbel MA, Philippe JM, Nguyen C, Victorero G, Freeman T, Wooding P, Miazek A, Mattei MG, Malissen M, Jordan BR, Malissen B, Carrier A, Naquet P (2000) The chemokine TECK is expressed by thymic and intestinal epithelial cells and attracts double- and single-positive thymocytes expressing the TECK receptor CCR9. Eur J Immunol 30(1):262–271
Staton TL, Habtezion A, Winslow MM, Sato T, Love PE, Butcher EC (2006) CD8+ recent thymic emigrants home to and efficiently repopulate the small intestine epithelium. Nat Immunol 7(5):482–488. https://doi.org/10.1038/ni1319
Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, Chen W (2008) A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat Immunol 9(6):632–640. https://doi.org/10.1038/ni.1607
Huehn J, Siegmund K, Lehmann JCU, Siewert C, Haubold U, Feuerer M, Debes GF, Lauber J, Frey O, Przybylski GK, Niesner U, de la Rosa M, Schmidt CA, Bauer R, Buer J, Scheffold A, Hamann A (2004) Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4(+) regulatory T cells. J Exp Med 199(3):303–313. https://doi.org/10.1084/jem.20031562
Sharma R, Sung SSJ, Abaya CE, Ju ACY, Fu SM, Ju ST (2009) IL-2 Regulates CD103 expression on CD4(+) T cells in scurfy mice that display both CD103-dependent and independent inflammation. J Immunol 183(2):1065–1073. https://doi.org/10.4049/jimmunol.0804354
Szabo PA, Miron M, Farber DL (2019) Location, location, location: tissue resident memory T cells in mice and humans. Sci Immunol. https://doi.org/10.1126/sciimmunol.aas9673
Park JY, DiPalma DT, Kwon J, Fink J, Park JH (2019) Quantitative difference in PLZF protein expression determines iNKT lineage fate and controls innate CD8 T cell generation. Cell Rep 27(9):2548–2557. https://doi.org/10.1016/j.celrep.2019.05.012
Zhang JH, Dong ZJ, Zhou RB, Luo DM, Wei HM, Tian ZG (2005) Isolation of lymphocytes and their innate immune characterizations from liver, intestine. Lung Uterus Cell Mol Immunol 2(4):271–280
Acknowledgements
We thank the General Surgery Department and Residency Program, Guthrie Robert Packer Hospital, for support in allowing Hilary R. Keller an interruption in her residency training to join the National Institutes of Health Cancer Research Training Award fellowship program.
Funding
This study has been supported by the Intramural Research Program of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research, and by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI18C0479).
Author information
Authors and Affiliations
Contributions
HRK, DLL, and CL designed and performed the experiments, analyzed the data, and contributed to the writing of the manuscript. SH, MAL, PP, NL, AC, and VL performed the experiments, analyzed the data, and commented on the manuscript. YKP and JHP conceived the project, analyzed the data, and wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
All authors consent to the publication of this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Keller, H.R., Ligons, D.L., Li, C. et al. The molecular basis and cellular effects of distinct CD103 expression on CD4 and CD8 T cells. Cell. Mol. Life Sci. 78, 5789–5805 (2021). https://doi.org/10.1007/s00018-021-03877-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-021-03877-9