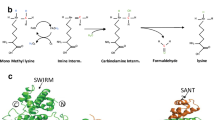
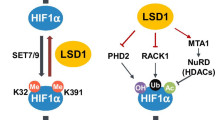
Abstract
It is well-established that Lysine-specific demethylase 1 (LSD1, also known as KDM1A) roles as a lysine demethylase canonically acting on H3K4me1/2 and H3K9me1/2 for regulating gene expression. Though the discovery of non-histone substrates methylated by LSD1 has largely expanded the functions of LSD1 as a typical demethylase, recent groundbreaking studies unveiled its non-catalytic functions as a second life for this demethylase. We and others found that LSD1 is implicated in the interaction with a line of proteins to exhibit additional non-canonical functions in a demethylase-independent manner. Here, we present an integrated overview of these recent literatures charging LSD1 with unforeseen functions to re-evaluate and summarize its non-catalytic biological roles beyond the current understanding of its demethylase activity. Given LSD1 is reported to be ubiquitously overexpressed in a variety of tumors, it has been generally considered as an innovative target for cancer therapy. We anticipate that these non-canonical functions of LSD1 will arouse the consideration that extending the LSD1-based drug discovery to targeting LSD1 protein interactions non-catalytically, not only its demethylase activity, may be a novel strategy for cancer prevention.



Similar content being viewed by others
Abbreviations
- AR:
-
Androgen receptor
- AML:
-
Acute myeloid leukemia
- AOL:
-
Amine oxidase-like
- CoREST:
-
REST (RE1-silencing transcription factor) corepressor
- CPD:
-
Cdc4 phosphodegron
- CREB:
-
CAMP-response element-binding protein
- CtBP:
-
C-terminal-binding protein 1
- Dnmt1:
-
DNA methyltransferase 1
- E2F1:
-
Transcription factor
- EMT:
-
Epithelial-to-mesenchymal transition
- ER:
-
Estrogen receptor
- ERRα:
-
Estrogen-related receptor α
- GFI:
-
Growth factor independence
- H3K4me1/2:
-
Mono- and di-methylated histone 3 lysine 4
- H3K9me1/2:
-
Mono- and di-methylated histone 3 lysine 9
- H4K20me2:
-
Di-methylated histone 4 lysine 20
- HIF-1α:
-
Hypoxia-inducible factor-1
- LSD1:
-
Lysine-specific demethylase 1
- MAO:
-
Monoamine oxidase
- MEF2:
-
Myocyte enhancer factor 2
- NHEJ:
-
Nonhomologous end-joining
- NuRD:
-
Nucleosome remodeling and deacetylase
- PTMs:
-
Post-translational modifications
- SCF:
-
SKP1-CUL1-F-box protein
- STAT3:
-
Signal transducer and activator of transcription 3
- SQSTM1:
-
Sequestasome 1
References
Shi Y et al (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119(7):941–953
Shi YJ et al (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19(6):857–864
Lee MG, Wynder C, Cooch N, Shiekhattar R (2005) An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437(7057):432–435
Laurent B et al (2015) A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol Cell 57(6):957–970
Metzger E et al (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437(7057):436–439
Wang J et al (2007) Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446(7138):882–887
Huang J et al (2007) p53 is regulated by the lysine demethylase LSD1. Nature 449(7158):105–108
He Y et al (2018) LSD1 promotes S-phase entry and tumorigenesis via chromatin co-occupation with E2F1 and selective H3K9 demethylation. Oncogene 37(4):534–543
Zhang J et al (2011) Cyclophosphamide perturbs cytosine methylation in Jurkat-T cells through LSD1-mediated stabilization of DNMT1 protein. Chem Res Toxicol 24(11):2040–2043
Lee JY et al (2017) LSD1 demethylates HIF1alpha to inhibit hydroxylation and ubiquitin-mediated degradation in tumor angiogenesis. Oncogene 36(39):5512–5521
Yang JB et al (2010) Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA 107(50):21499–21504
Lan F, Nottke AC, Shi Y (2008) Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol 20(3):316–325
Ambrosio S, Sacca CD, Majello B (2017) Epigenetic regulation of epithelial to mesenchymal transition by the Lysine-specific demethylase LSD1/KDM1A. Biochim Biophys Acta Gene Regul Mech 1860(9):905–910
Lv S et al (2010) LSD1 is required for chromosome segregation during mitosis. Eur J Cell Biol 89(7):557–563
Sakamoto A et al (2015) Lysine demethylase LSD1 coordinates glycolytic and mitochondrial metabolism in hepatocellular carcinoma cells. Cancer Res 75(7):1445–1456
Whyte WA et al (2012) Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482(7384):221–225
Amente S, Lania L, Majello B (2013) The histone LSD1 demethylase in stemness and cancer transcription programs. Biochim Biophys Acta 1829(10):981–986
Lan H et al (2019) LSD1 destabilizes FBXW7 and abrogates FBXW7 functions independent of its demethylase activity. Proc Natl Acad Sci USA 116(25):12311–12320
Chao A et al (2017) Lysine-specific demethylase 1 (LSD1) destabilizes p62 and inhibits autophagy in gynecologic malignancies. Oncotarget 8(43):74434–74450
Si W, Zhao Y, Zhou J, Zhang Q, Zhang Y (2019) The coordination between ZNF217 and LSD1 contributes to hepatocellular carcinoma progress and is negatively regulated by miR-101. Exp Cell Res 379(1):1–10
Aravind L, Iyer LM (2002) The SWIRM domain: a conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol 3(8):reserch0039-1
Chen Y et al (2006) Crystal structure of human histone lysine-specific demethylase 1 (LSD1). P Natl Acad Sci USA 103(38):13956–13961
Niwa H, Umehara T (2017) Structural insight into inhibitors of flavin adenine dinucleotide-dependent lysine demethylases. Epigenetics 12(5):340–352
Fang R et al (2013) LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Mol Cell 49(3):558–570
Zhang Q et al (2013) Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b. Cell Re 23(2):225–241
Peng B et al (2015) Modulation of LSD1 phosphorylation by CK2/WIP1 regulates RNF168-dependent 53BP1 recruitment in response to DNA damage. Nucleic Acids Res 43(12):5936–5947
Metzger E et al (2016) Assembly of methylated KDM1A and CHD1 drives androgen receptor-dependent transcription and translocation. Nat Struct Mol Biol 23(2):132–139
Stavropoulos P, Blobel G, Hoelz A (2006) Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol 13(7):626–632
Shi Y et al (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422(6933):735–738
Wang Y et al (2009) LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138(4):660–672
Perillo B et al (2008) DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319(5860):202–206
Cai CM et al (2014) Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep 9(5):1618–1627
Wang J et al (2015) LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat Neurosci 18(9):1256–1264
Hojfeldt JW, Agger K, Helin K (2013) Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov 12(12):917–930
Saleque S, Kim JW, Rooke HM, Orkin SH (2007) Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell 27(4):562–572
Wu ZQ et al (2012) Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci USA 109(41):16654–16659
Lin YW et al (2010) The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. Embo J 29(11):1803–1816
Ferrari-Amorotti G et al (2013) Inhibiting interactions of lysine demethylase LSD1 with snail/slug blocks cancer cell invasion. Cancer Res 73(1):235–245
Egolf S et al (2019) LSD1 inhibition promotes epithelial differentiation through derepression of fate-determining transcription factors. Cell Rep 28(8):1981
Maiques-Diaz A et al (2018) Enhancer activation by pharmacologic displacement of LSD1 from GFI1 induces differentiation in acute myeloid leukemia. Cell Rep 22(13):3641–3659
Majello B, Gorini F, Sacca CD, Amente S (2019) Expanding the role of the histone lysine-specific demethylase LSD1 in cancer. Cancers 11(3):324
Shi Y (2007) Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet 8(11):829–833
Zheng YC et al (2015) A Systematic review of histone lysine-specific demethylase 1 and its inhibitors. Med Res Rev 35(5):1032–1071
Cho HS et al (2011) Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res 71(3):655–660
Kontaki H, Talianidis I (2010) Lysine methylation regulates E2F1-induced cell death. Mol Cell 39(1):152–160
Baek SH, Kim KI (2016) Regulation of HIF-1 alpha stability by lysine methylation. Bmb Rep 49(5):245–246
Sheng W et al (2018) LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174(3):549–563e519
Lan H et al (2019) LSD1 destabilizes FBXW7 and abrogates FBXW7 functions independent of its demethylase activity. Proc Natl Acad Sci USA 116:12311–12320
Welcker M, Clurman BE (2008) FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 8(2):83–93
Wang Z, Liu P, Inuzuka H, Wei W (2014) Roles of F-box proteins in cancer. Nat Rev Cancer 14(4):233–247
Horard B, Vanacker JM (2003) Estrogen receptor-related receptors: orphan receptors desperately seeking a ligand. J Mol Endocrinol 31(3):349–357
Carnesecchi J et al (2017) ERRalpha induces H3K9 demethylation by LSD1 to promote cell invasion. Proc Natl Acad Sci USA 114(15):3909–3914
Carnesecchi J, Cerutti C, Vanacker JM, Forcet C (2017) ERR alpha protein is stabilized by LSD1 in a demethylation-independent manner. PLoS One 12(11):e0188871
Moscat J, Diaz-Meco MT (2009) p62 at the Crossroads of Autophagy, Apoptosis, and Cancer. Cell 137(6):1001–1004
Moscat J, Diaz-Meco MT (2012) p62: a versatile multitasker takes on cancer. Trends Biochem Sci 37(6):230–236
Green SM, Mostaghel EA, Nelson PS (2012) Androgen action and metabolism in prostate cancer. Mol Cell Endocrinol 360(1–2):3–13
Yuan X et al (2014) Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene 33(22):2815–2825
Sehrawat A et al (2018) LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Cancer Res 78(13):E4179–E4188
Mohammad HP et al (2015) A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 28(1):57–69
McGrath JP et al (2016) Pharmacological inhibition of the histone lysine demethylase KDM1A suppresses the growth of multiple acute myeloid leukemia subtypes. Cancer Res 76(7):1975–1988
Fiskus W et al (2014) Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia 28(11):2155–2164
Theisen ER et al (2014) Reversible inhibition of lysine specific demethylase 1 is a novel anti-tumor strategy for poorly differentiated endometrial carcinoma. Bmc Cancer 14:752
Kahl P et al (2006) Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res 66(23):11341–11347
Schulte JH et al (2009) Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res 69(5):2065–2071
Kauffman EC et al (2011) Role of androgen receptor and associated lysine-demethylase coregulators, LSD1 and JMJD2A, in localized and advanced human bladder cancer. Mol Carcinog 50(12):931–944
Harris WJ et al (2012) The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21(4):473–487
Lv T et al (2012) Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS ONE 7(4):e35065
Fu X, Zhang P, Yu B (2017) Advances toward LSD1 inhibitors for cancer therapy. Future Med Chem 9(11):1227–1242
Hosseini A, Minucci S (2017) A comprehensive review of lysine-specific demethylase 1 and its roles in cancer. Epigenomics 9(8):1123–1142
Deshaies RJ (2015) Protein degradation: prime time for PROTACs. Nat Chem Biol 11(9):634–635
Lan H, Sun Y (2019) FBXW7 E3 ubiquitin ligase: degrading, not degrading, or being degraded. Protein Cell 10:861–863
Wang J et al (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41(1):125–129
Zhang X et al (2013) Regulation of estrogen receptor alpha by histone methyltransferase SMYD2-mediated protein methylation. Proc Natl Acad Sci USA 110(43):17284–17289
Nair SS, Li DQ, Kumar R (2013) A core chromatin remodeling factor instructs global chromatin signaling through multivalent reading of nucleosome codes. Mol Cell 49(4):704–718
Acknowledgements
This study was supported in part by the Natural Science Foundation of Zhejiang Province (20160171) (YZW), by the Ningbo Natural Science Foundation, China Grant (No. 2016A610148) (ZYY), and by the Medical Scientific Research Foundation of Zhejiang Province, China (Grant No. 2018KY689) (ZYY). Authors’ contributions: HYL had the idea for the article and providing the final approval of the version to be published. YXL was involved in performed the literature search and revision work, FYG was involved in drafting the manuscript. XXW and ZYY were involved in checking the language problems. YZW was involved in revising the manuscript critically for important scientific content. And the authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gu, F., Lin, Y., Wang, Z. et al. Biological roles of LSD1 beyond its demethylase activity. Cell. Mol. Life Sci. 77, 3341–3350 (2020). https://doi.org/10.1007/s00018-020-03489-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-020-03489-9