Abstract
The UDP-glucose ceramide glucosyltransferase (UGCG) is a key enzyme in the synthesis of glycosylated sphingolipids, since this enzyme generates the precursor for all complex glycosphingolipids (GSL), the GlcCer. The UGCG has been associated with several cancer-related processes such as maintaining cancer stem cell properties or multidrug resistance induction. The precise mechanisms underlying these processes are unknown. Here, we investigated the molecular mechanisms occurring after UGCG overexpression in breast cancer cells. We observed alterations of several cellular properties such as morphological changes, which enhanced proliferation and doxorubicin resistance in UGCG overexpressing MCF-7 cells. These cellular effects seem to be mediated by an altered composition of glycosphingolipid-enriched microdomains (GEMs), especially an accumulation of globotriaosylceramide (Gb3) and glucosylceramide (GlcCer), which leads to an activation of Akt and ERK1/2. The induction of the Akt and ERK1/2 signaling pathway results in an increased gene expression of multidrug resistance protein 1 (MDR1) and anti-apoptotic genes and a decrease of pro-apoptotic gene expression. Inhibition of the protein kinase C (PKC) and phosphoinositide 3 kinase (PI3K) reduced MDR1 gene expression. This study discloses how changes in UGCG expression impact several cellular signaling pathways in breast cancer cells resulting in enhanced proliferation and multidrug resistance.







Similar content being viewed by others
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359–E386
Ichikawa S, Sakiyama H, Suzuki G, Hidari KI, Hirabayashi Y (1996) Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc Natl Acad Sci USA 93:12654
Wegner MS, Schiffmann S, Parnham MJ, Geisslinger G, Grosch S (2016) The enigma of ceramide synthase regulation in mammalian cells. Prog Lipid Res 63:93–119
Yamashita T, Wada R, Sasaki T, Deng C, Bierfreund U, Sandhoff K, Proia RL (1999) A vital role for glycosphingolipid synthesis during development and differentiation. Proc Natl Acad Sci USA 96:9142–9147
Jennemann R, Sandhoff R, Langbein L, Kaden S, Rothermel U, Gallala H, Sandhoff K, Wiegandt H, Grone HJ (2007) Integrity and barrier function of the epidermis critically depend on glucosylceramide synthesis. J Biol Chem 282:3083–3094
Amen N, Mathow D, Rabionet M, Sandhoff R, Langbein L, Gretz N, Jackel C, Grone HJ, Jennemann R (2013) Differentiation of epidermal keratinocytes is dependent on glucosylceramide: ceramide processing. Hum Mol Genet 22:4164–4179
Allende ML, Proia RL (2014) Simplifying complexity: genetically resculpting glycosphingolipid synthesis pathways in mice to reveal function. Glycoconj J 31:613–622
Mistry PK, Lukina E, Ben Turkia H, Shankar SP, Baris H, Ghosn M, Mehta A, Packman S, Pastores G, Petakov M, Assouline S, Balwani M, Danda S, Hadjiev E, Ortega A, Gaemers SJM, Tayag R, Peterschmitt MJ (2017) Outcomes after 18 months of eliglustat therapy in treatment-naive adults with gaucher disease type 1: the phase 3 ENGAGE trial. Am J Hematol 92:1170–1176
Gouaze V, Yu JY, Bleicher RJ, Han TY, Liu YY, Wang H, Gottesman MM, Bitterman A, Giuliano AE, Cabot MC (2004) Overexpression of glucosylceramide synthase and P-glycoprotein in cancer cells selected for resistance to natural product chemotherapy. Mol Cancer Ther 3:633–639
Gouaze-Andersson V, Yu JY, Kreitenberg AJ, Bielawska A, Giuliano AE, Cabot MC (2007) Ceramide and glucosylceramide upregulate expression of the multidrug resistance gene MDR1 in cancer cells. Biochim Biophys Acta 1771:1407–1417
Jennemann R, Grone HJ (2013) Cell-specific in vivo functions of glycosphingolipids: lessons from genetic deletions of enzymes involved in glycosphingolipid synthesis. Prog Lipid Res 52:231–248
Ishibashi Y, Kohyama-Koganeya A, Hirabayashi Y (2013) New insights on glucosylated lipids: metabolism and functions. Biochim Biophys Acta 1831:1475–1485
Liu YY, Hill RA, Li YT (2013) Ceramide glycosylation catalyzed by glucosylceramide synthase and cancer drug resistance. Adv Cancer Res 117:59–89
Liu YY, Gupta V, Patwardhan GA, Bhinge K, Zhao Y, Bao J, Mehendale H, Cabot MC, Li YT, Jazwinski SM (2010) Glucosylceramide synthase upregulates MDR1 expression in the regulation of cancer drug resistance through cSrc and beta-catenin signaling. Mol Cancer 9:145
Morad SA, Cabot MC (2015) Tamoxifen regulation of sphingolipid metabolism—therapeutic implications. Biochim Biophys Acta 1851:1134–1145
Liu J, Zhang X, Liu A, Zhang D, Su Y, Liu Y, You D, Yuan L, Kong X, Wang X, Sun P (2016) Altered methylation of glucosylceramide synthase promoter regulates its expression and associates with acquired multidrug resistance in invasive ductal breast cancer. Oncotarget 7(24):36755–36766
Zhang X, Wu X, Su P, Gao Y, Meng B, Sun Y, Li L, Zhou Z, Zhou G (2012) Doxorubicin influences the expression of glucosylceramide synthase in invasive ductal breast cancer. PLoS One 7:e48492
Crespo PM, Silvestre DC, Gil GA, Maccioni HJ, Daniotti JL, Caputto BL (2008) c-Fos activates glucosylceramide synthase and glycolipid synthesis in PC12 cells. J Biol Chem 283:31163–31171
Di Sano F, Fazi B, Citro G, Lovat PE, Cesareni G, Piacentini M (2003) Glucosylceramide synthase and its functional interaction with RTN-1C regulate chemotherapeutic-induced apoptosis in neuroepithelioma cells. Cancer Res 63:3860–3865
Mollinedo F, Gajate C (2015) Lipid rafts as major platforms for signaling regulation in cancer. Adv Biol Regul 57:130–146
Staubach S, Hanisch FG (2011) Lipid rafts: signaling and sorting platforms of cells and their roles in cancer. Expert Rev Proteom 8:263–277
Yamaji T, Horie A, Tachida Y, Sakuma C, Suzuki Y, Kushi Y, Hanada K (2016) Role of intracellular lipid logistics in the preferential usage of very long chain-ceramides in glucosylceramide. Int J Mol Sci 17:1761
Newton J, Lima S, Maceyka M, Spiegel S (2015) Revisiting the sphingolipid rheostat: evolving concepts in cancer therapy. Exp Cell Res 333:195–200
Tornquist K (2013) Sphingosine 1-phosphate and cancer: lessons from thyroid cancer cells. Biomolecules 3:303–315
Hakomori SI (2000) Cell adhesion/recognition and signal transduction through glycosphingolipid microdomain. Glycoconj J 17:143–151
Sabharanjak S, Sharma P, Parton RG, Mayor S (2002) GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev Cell 2:411–423
Rogers KR, Kikawa KD, Mouradian M, Hernandez K, McKinnon KM, Ahwah SM, Pardini RS (2010) Docosahexaenoic acid alters epidermal growth factor receptor-related signaling by disrupting its lipid raft association. Carcinogenesis 31:1523–1530
Hers I, Vincent EE, Tavare JM (2011) Akt signalling in health and disease. Cell Signal 23:1515–1527
Wee P, Wang Z (2017) Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 9:52
Hartmann D, Wegner MS, Wanger RA, Ferreiros N, Schreiber Y, Lucks J, Schiffmann S, Geisslinger G, Grosch S (2013) The equilibrium between long and very long chain ceramides is important for the fate of the cell and can be influenced by co-expression of CerS. Int J Biochem Cell Biol 45:1195–1203
Mesicek J, Lee H, Feldman T, Jiang X, Skobeleva A, Berdyshev EV, Haimovitz-Friedman A, Fuks Z, Kolesnick R (2010) Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cell Signal 22:1300–1307
Yamane M, Miyazawa K, Moriya S, Abe A, Yamane S (2011) D, L-Threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol (DL-PDMP) increases endoplasmic reticulum stress, autophagy and apoptosis accompanying ceramide accumulation via ceramide synthase 5 protein expression in A549 cells. Biochimie 93:1446–1459
Payne AW, Pant DK, Pan TC, Chodosh LA (2014) Ceramide kinase promotes tumor cell survival and mammary tumor recurrence. Cancer Res 74:6352–6363
Burns TA, Subathra M, Signorelli P, Choi Y, Yang X, Wang Y, Villani M, Bhalla K, Zhou D, Luberto C (2013) Sphingomyelin synthase 1 activity is regulated by the BCR–ABL oncogene. J Lipid Res 54:794–805
Separovic D, Semaan L, Tarca AL, Awad Maitah MY, Hanada K, Bielawski J, Villani M, Luberto C (2008) Suppression of sphingomyelin synthase 1 by small interference RNA is associated with enhanced ceramide production and apoptosis after photodamage. Exp Cell Res 314:1860–1868
Jin ZX, Huang CR, Dong L, Goda S, Kawanami T, Sawaki T, Sakai T, Tong XP, Masaki Y, Fukushima T, Tanaka M, Mimori T, Tojo H, Bloom ET, Okazaki T, Umehara H (2008) Impaired TCR signaling through dysfunction of lipid rafts in sphingomyelin synthase 1 (SMS1)-knockdown T cells. Int Immunol 20:1427–1437
Hartmann D, Lucks J, Fuchs S, Schiffmann S, Schreiber Y, Ferreiros N, Merkens J, Marschalek R, Geisslinger G, Grosch S (2012) Long chain ceramides and very long chain ceramides have opposite effects on human breast and colon cancer cell growth. Int J Biochem Cell Biol 44:620–628
He X, Schuchman EH (2012) Potential role of acid sphingomyelinase in environmental health. Zhong nan da xue xue bao Yi xue ban J Cent South Univ Med Sci 37:109–125
Yabu T, Shiba H, Shibasaki Y, Nakanishi T, Imamura S, Touhata K, Yamashita M (2015) Stress-induced ceramide generation and apoptosis via the phosphorylation and activation of nSMase1 by JNK signaling. Cell Death Differ 22:258–273
Park B, Lee YM, Kim JS, Her Y, Kang JH, Oh SH, Kim HM (2013) Neutral sphingomyelinase 2 modulates cytotoxic effects of protopanaxadiol on different human cancer cells. BMC Complement Altern Med 13:194
Kim WJ, Okimoto RA, Purton LE, Goodwin M, Haserlat SM, Dayyani F, Sweetser DA, McClatchey AI, Bernard OA, Look AT, Bell DW, Scadden DT, Haber DA (2008) Mutations in the neutral sphingomyelinase gene SMPD3 implicate the ceramide pathway in human leukemias. Blood 111:4716–4722
Marchesini N, Luberto C, Hannun YA (2003) Biochemical properties of mammalian neutral sphingomyelinase 2 and its role in sphingolipid metabolism. J Biol Chem 278:13775–13783
Sanger N, Ruckhaberle E, Gyorffy B, Engels K, Heinrich T, Fehm T, Graf A, Holtrich U, Becker S, Karn T (2015) Acid ceramidase is associated with an improved prognosis in both DCIS and invasive breast cancer. Mol Oncol 9:58–67
Sukocheva O, Wadham C, Gamble J, Xia P (2015) Sphingosine-1-phosphate receptor 1 transmits estrogens’ effects in endothelial cells. Steroids 104:237–245
Shen W, Henry AG, Paumier KL, Li L, Mou K, Dunlop J, Berger Z, Hirst WD (2014) Inhibition of glucosylceramide synthase stimulates autophagy flux in neurons. J Neurochem 129:884–894
Gosejacob D, Jager PS, Vom Dorp K, Frejno M, Carstensen AC, Kohnke M, Degen J, Dormann P, Hoch M (2016) Ceramide synthase 5 is essential to maintain C16:0-ceramide pools and contributes to the development of diet-induced obesity. J Biol Chem 291:6989–7003
White MD, Chan L, Antoon JW, Beckman BS (2013) Targeting ovarian cancer and chemoresistance through selective inhibition of sphingosine kinase-2 with ABC294640. Anticancer Res 33:3573–3579
Katayama K, Noguchi K, Sugimoto Y (2014) Regulations of P-glycoprotein/ABCB1/MDR1 in human cancer cells. New J Sci 2014:10
Dahdouh F, Raane M, Thevenod F, Lee WK (2014) Nickel-induced cell death and survival pathways in cultured renal proximal tubule cells: roles of reactive oxygen species, ceramide and ABCB1. Arch Toxicol 88:881–892
Sharom FJ (2014) Complex interplay between the P-glycoprotein multidrug efflux pump and the membrane: its role in modulating protein function. Front Oncol 4:41
Bentires-Alj M, Barbu V, Fillet M, Chariot A, Relic B, Jacobs N, Gielen J, Merville MP, Bours V (2003) NF-kappaB transcription factor induces drug resistance through MDR1 expression in cancer cells. Oncogene 22:90–97
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Peng B, Weintraub ST, Coman C, Ponnaiyan S, Sharma R, Tews B, Winter D, Ahrends R (2017) A comprehensive high-resolution targeted workflow for the deep profiling of sphingolipids. Anal Chem 89:12480–12487
Acknowledgements
This work was funded by the Deutsche Forschungsgemeinschaft (WE 5825/1-1), SFB 1039 TP B05, the August Scheidel-Stiftung, the Heinrich und Fritz Riese-Stiftung, and Minerva-Stiftung. The supports by the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen, the Senatsverwaltung für Wirtschaft, Technologie und Forschung des Landes Berlin, and the Bundesministerium für Bildung und Forschung and BMBF (Code 031L0108A, 031A534B) are also gratefully acknowledged. In addition, funding by the Sigrid Jusélius Foundation and Magnus Ehrnrooth Foundation and Åbo Akademi University are acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2018_2799_MOESM1_ESM.pptx
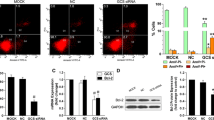
Supplementary material 1 (PPTX 553 kb) Supplemental 1 Stable UGCG knockdown in MCF-7 cells by CRISPR/Cas. (A) Validation of the UGCG knockdown by qRT-PCR, Western blot analysis and immunocytochemistry. Data are represented as a mean of n = 4 to 11 ± SEM. Unpaired t test with Welch’s correction. (B) Transmitted light image acquisition show a reduced cytoplasm size in MCF-7/UGCG KD cells as compared to control and MCF-7/UGCG OE cells. Representative images. (C) Determination of the relative living cell number of MCF-7 cells. Data are represented as a mean of n = 7 to 15 ± SEM. Unpaired t test with Welch’s correction. p ≤ * 0.05, p ≤ ** 0.01, p ≤ *** 0.001, p ≤ **** 0.0001
18_2018_2799_MOESM2_ESM.pptx
Supplementary material 2 (PPTX 489 kb) Supplemental 2 Cytoskeleton, cytoplasm staining and MDR1 mRNA expression of MCF-7 cells. (A) Immunocytochemistry of MCF-7 cells. Cytoskeleton is indicated as β-actin staining (upper panel). Cell membrane is indicated as β-catenin staining (lower panel). Nuclei are stained by DAPI. (B) Analysis of MDR1 mRNA expression by qRT-PCR. The expression is related to the housekeeping gene RPL37A. Data are represented as a mean of n = 3-6 ± SEM. Unpaired t test with Welch’s correction. p ≤ *** 0.001
18_2018_2799_MOESM3_ESM.pptx
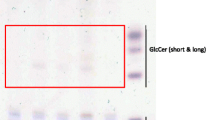
Supplementary material 3 (PPTX 487 kb) Supplemental 3 Analysis of mRNA expression of sphingolipid enzymes by qRT-PCR and sphingolipid concentration determination by LC–MS/MS in 3D MCF-7 spheroids. (A) Analysis of sphingolipid enzymes mRNA expression by qRT-PCR. The expression is related to the housekeeping gene RPL37A. Data are represented as a mean of n = 3 ± SEM. (B) Concentrations of C14:0-, C16:0-, C18:0-, C18:1-, C20:0-, C24:0-Cer determined by LC–MS/MS. (C) Concentrations of C16:0-, C18:0-, C18:1-, C24:1-GlcCer, C16:0-, C18:0-, C24:0- and C24:1-LacCer determined by LC–MS/MS. (D) Total Cer, Glc- and LacCer levels determined by LC–MS/MS. Unpaired t test with Welch’s correction. p ≤ * 0.05, p ≤ ** 0.01, p ≤ *** 0.001, p ≤ **** 0.0001
18_2018_2799_MOESM4_ESM.pptx
Supplementary material 4 (PPTX 16050 kb) Supplemental 4 Analysis of the size and nuclei staining of 3D MCF-7 spheroids by microscopy. The transmitted light microscopy shows that MCF-7/UGCG OE spheroids are more densely packed than MCF-7/naiv and MCF-7/pTarget cells. DAPI staining indicates cell nuclei. Representative images
18_2018_2799_MOESM5_ESM.pptx
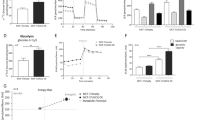
Supplementary material 5 (PPTX 283 kb) Supplemental 5 Determination of sphingolipid concentrations in MCF-7 cells by LC–MS/MS. (A) Sph-1p concentrations in MCF-7/pTarget and MCF-7/UGCG OE cells following 0 and 2 µM PPMP stimulation. (B) GEMs were isolated and total sphinganine, Cer and LacCer levels of fractions 1 to 10 were determined by LC–MS/MS. Data are represented as a mean of n = 3 ± SEM. p ≤ * 0.05
18_2018_2799_MOESM6_ESM.pptx
Supplementary material 6 (PPTX 442 kb) Supplemental 6 Analysis of different cellular marker in fractions 1 to 10 in MCF-7 cells by Western blot analysis. (A) PonceauS staining. (B) Western blot analysis of different cellular marker
18_2018_2799_MOESM7_ESM.pptx
Supplementary material 7 (PPTX 427 kb) Supplemental 7 Analysis of phosphorylation status of key signaling molecules in MCF-7 cells following PPMP stimulation by an antibody-based array. Array was performed according to manufacturer’s protocol. (A) Densitometrical analysis of the array. Data are represented as a mean of n = 2 ± SEM. RFU = relative fluorescence unit. (B) Representative arrays were displayed. p ≤ * 0.05, p ≤ ** 0.01, p ≤ **** 0.0001
18_2018_2799_MOESM8_ESM.pptx
Supplementary material 8 (PPTX 427 kb) Supplemental 8 Analysis of phosphorylation status of key signaling molecules in 3D MCF-7 spheroids by an antibody-based array. Array was performed according to manufacturer’s protocol. (A) Densitometrical analysis of the array. Data are represented as a mean of n = 4 ± SEM. (B) Representative arrays were displayed. RFU = relative fluorescence unit. RFU = relative fluorescence unit. p ≤ * 0.05, p ≤ **** 0.0001
18_2018_2799_MOESM9_ESM.pptx
Supplementary material 9 (PPTX 588 kb) Supplemental 9 Analysis of phosphorylation status of key signaling molecules in MCF-7/UGCG KD cells by an antibody-based array. Array was performed according to manufacturer’s protocol. (A) Densitometrical analysis of the array. Data are represented as a mean of n = 5-7 ± SEM. RFU = relative fluorescence unit. (B) Representative arrays were displayed. p ≤ * 0.05
Rights and permissions
About this article
Cite this article
Wegner, MS., Schömel, N., Gruber, L. et al. UDP-glucose ceramide glucosyltransferase activates AKT, promoted proliferation, and doxorubicin resistance in breast cancer cells. Cell. Mol. Life Sci. 75, 3393–3410 (2018). https://doi.org/10.1007/s00018-018-2799-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2799-7