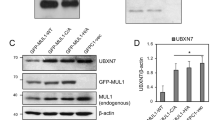
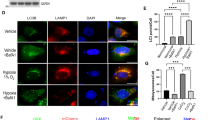
Abstract
Cell stress such as hypoxia elicits adaptive responses, also on the level of mitochondria, and in part is mediated by the hypoxia-inducible factor (HIF) 1α. Adaptation of mitochondria towards acute hypoxic conditions is reasonably well understood, while regulatory mechanisms, especially of respiratory chain assembly factors, under chronic hypoxia remains elusive. One of these assembly factors is transmembrane protein 126B (TMEM126B). This protein is part of the mitochondrial complex I assembly machinery. We identified changes in complex I abundance under chronic hypoxia, in association with impaired substrate-specific mitochondrial respiration. Complexome profiling of isolated mitochondria of the human leukemia monocytic cell line THP-1 revealed HIF-1α-dependent deficits in complex I assembly and mitochondrial complex I assembly complex (MCIA) abundance. Of all mitochondrial MCIA members, we proved a selective HIF-1-dependent decrease of TMEM126B under chronic hypoxia. Mechanistically, HIF-1α induces the E3-ubiquitin ligase F-box/WD repeat-containing protein 1A (β-TrCP1), which in turn facilitates the proteolytic degradation of TMEM126B. Attenuating a functional complex I assembly appears critical for cellular adaptation towards chronic hypoxia and is linked to destruction of the mitochondrial assembly factor TMEM126B.






Similar content being viewed by others
Abbreviations
- ACAD9:
-
Acyl-CoA dehydrogenase family member 9
- ADP:
-
Adenosine diphosphate
- Akt:
-
RAC-alpha serine/threonine-protein kinase
- ATP:
-
Adenosine triphosphate
- ATP5a:
-
ATP synthase subunit alpha
- β-TrCP:
-
F-box/WD repeat-containing protein 1A
- BNE:
-
Blue native electrophoresis
- ChIP:
-
Chromatin immunoprecipitation
- CHX:
-
Cycloheximide
- COX:
-
Cytochrome c oxidase subunit
- DMOG:
-
Dimethyloxalylglycine
- DMSO:
-
Dimethyl sulfoxide
- ECSIT:
-
Evolutionarily conserved signaling intermediate in toll pathway
- ETF:
-
Electron transfer flavoprotein
- FAD:
-
Flavin adenine dinucleotide
- FCCP:
-
Carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone
- GFP:
-
Green fluorescent protein
- GPDH:
-
Glycerol-3-phosphate dehydrogenase
- GSK3:
-
Glycogen synthase kinase-3
- HIF:
-
Hypoxia-inducible factor
- HIGD:
-
Hypoxia-inducible gene
- IP:
-
Immunoprecipitation
- IPTG:
-
Isopropyl-β-d-thiogalactopyranoside
- KCN:
-
Potassium cyanide
- LC:
-
Lactacystin
- MCIA:
-
Mitochondrial complex I assembly complex
- NAD:
-
Nicotinamide adenine dinucleotide
- ND:
-
NADH-ubiquinone oxidoreductase chain
- NDUFA:
-
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit
- NDUFAF1:
-
Complex I intermediate-associated protein 30
- NDUFA4L2:
-
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 4-like 2
- NDUFB:
-
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit
- NDUFS:
-
NADH dehydrogenase [ubiquinone] iron–sulfur protein
- OXPHOS:
-
Oxidative phosphorylation
- PHD:
-
Prolyl hydroxylase
- PI3K:
-
Phosphatidylinositol 3-kinase
- rot:
-
Rotenone
- shC:
-
Cells transduced with control short hairpin RNA
- sh1:
-
Cells transduced with short hairpin RNA against HIF-1α
- sh2:
-
Cells transduced with short hairpin RNA against HIF-2α
- TIMMDC1:
-
Translocase of inner mitochondrial membrane domain-containing protein 1
- TMEM126B:
-
Transmembrane protein 126B
References
Scholz CC, Taylor CT (2013) Targeting the HIF pathway in inflammation and immunity. Curr Opin Pharmacol 13(4):646–653. https://doi.org/10.1016/j.coph.2013.04.009
Palazon A, Goldrath AW, Nizet V et al (2014) HIF transcription factors, inflammation, and immunity. Immunity 41(4):518–528. https://doi.org/10.1016/j.immuni.2014.09.008
Mucaj V, Shay JE, Simon MC (2012) Effects of hypoxia and HIFs on cancer metabolism. Int J Hematol 95(5):464–470. https://doi.org/10.1007/s12185-012-1070-5
Catrina S-B, Okamoto K, Pereira T et al (2004) Hyperglycemia regulates hypoxia-inducible factor-1 protein stability and function. Diabetes 53(12):3226–3232. https://doi.org/10.2337/diabetes.53.12.3226
Bayer C, Shi K, Astner ST et al (2011) Acute versus chronic hypoxia. Why a simplified classification is simply not enough. Int J Radiat Oncol Biol Phys 80(4):965–968. https://doi.org/10.1016/j.ijrobp.2011.02.049
Bayer C, Vaupel P (2012) Acute versus chronic hypoxia in tumors. Controversial data concerning time frames and biological consequences. Strahlenther Onkol 188(7):616–627. https://doi.org/10.1007/s00066-012-0085-4
Fuhrmann DC, Wittig I, Heide H et al (1834) Chronic hypoxia alters mitochondrial composition in human macrophages. Biochim Biophys Acta 12:2750–2760. https://doi.org/10.1016/j.bbapap.2013.09.023
Huang LE, Gu J, Schau M et al (1998) Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA 95(14):7987–7992
Pugh CW, O’Rourke JF, Nagao M et al (1997) Activation of hypoxia-inducible factor-1; definition of regulatory domains within the alpha subunit. J Biol Chem 272(17):11205–11214
Appelhoff RJ, Tian YM, Raval RR et al (2004) Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem 279(37):38458–38465. https://doi.org/10.1074/jbc.M406026200
Jiang BH, Zheng JZ, Leung SW et al (1997) Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. Modulation of transcriptional activity by oxygen tension. J Biol Chem 272(31):19253–19260
Berchner-Pfannschmidt U, Tug S, Kirsch M et al (2010) Oxygen-sensing under the influence of nitric oxide. Cell Signal 22(3):349–356. https://doi.org/10.1016/j.cellsig.2009.10.004
Kapitsinou PP, Rajendran G, Astleford L et al (2016) The endothelial PHD2/HIF-2 axis regulates pulmonary artery pressure in mice. Mol Cell Biol. https://doi.org/10.1128/MCB.01055-15
Fuhrmann DC, Tausendschon M, Wittig I et al (2015) Inactivation of tristetraprolin in chronic hypoxia provokes the expression of cathepsin B. Mol Cell Biol 35(3):619–630. https://doi.org/10.1128/mcb.01034-14
Hatefi Y (1985) The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem 54:1015–1069. https://doi.org/10.1146/annurev.bi.54.070185.005055
Brandt U (2006) Energy converting NADH. Quinone oxidoreductase (complex I). Annu Rev Biochem 75:69–92. https://doi.org/10.1146/annurev.biochem.75.103004.142539
Janssen RJ, Nijtmans LG, van den Heuvel LP et al (2006) Mitochondrial complex I. Structure, function and pathology. J Inherit Metab Dis 29(4):499–515. https://doi.org/10.1007/s10545-006-0362-4
Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R et al (2013) Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340(6140):1567–1570. https://doi.org/10.1126/science.1230381
Lee I, Bender E, Kadenbach B (2002) Control of mitochondrial membrane potential and ROS formation by reversible phosphorylation of cytochrome c oxidase. Mol Cell Biochem 234–235(1–2):63–70
Fuhrmann DC, Brune B (2017) Mitochondrial composition and function under the control of hypoxia. Redox Biol 12:208–215. https://doi.org/10.1016/j.redox.2017.02.012
Semenza GL (2007) Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J 405(1):1–9. https://doi.org/10.1042/bj20070389
Tello D, Balsa E, Acosta-Iborra B et al (2011) Induction of the mitochondrial NDUFA4L2 protein by HIF-1alpha decreases oxygen consumption by inhibiting complex I activity. Cell Metab 14(6):768–779. https://doi.org/10.1016/j.cmet.2011.10.008
Lai RK, Xu IM, Chiu DK et al (2016) NDUFA4L2 fine-tunes oxidative stress in hepatocellular carcinoma. Clin Cancer Res 22(12):3105–3117. https://doi.org/10.1158/1078-0432.ccr-15-1987
Li J, Bai C, Guo J et al (2017) NDUFA4L2 protects against ischaemia/reperfusion-induced cardiomyocyte apoptosis and mitochondrial dysfunction by inhibiting complex I. Clin Exp Pharmacol Physiol 44(7):779–786. https://doi.org/10.1111/1440-1681.12768
Hernansanz-Agustín P, Ramos E, Navarro E et al (2017) Mitochondrial complex I deactivation is related to superoxide production in acute hypoxia. Redox Biol 12:1040–1051. https://doi.org/10.1016/j.redox.2017.04.025
Papandreou I, Cairns RA, Fontana L et al (2006) HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3(3):187–197. https://doi.org/10.1016/j.cmet.2006.01.012
Schagger H, de Coo R, Bauer MF et al (2004) Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J Biol Chem 279(35):36349–36353. https://doi.org/10.1074/jbc.M404033200
Acin-Perez R, Fernandez-Silva P, Peleato ML et al (2008) Respiratory active mitochondrial supercomplexes. Mol Cell 32(4):529–539. https://doi.org/10.1016/j.molcel.2008.10.021
Maranzana E, Barbero G, Falasca AI et al (2013) Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid Redox Signal 19(13):1469–1480. https://doi.org/10.1089/ars.2012.4845
Schagger H, Pfeiffer K (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19(8):1777–1783. https://doi.org/10.1093/emboj/19.8.1777
Genova ML, Lenaz G (2014) Functional role of mitochondrial respiratory supercomplexes. Biochim Biophys Acta 4:427–443. https://doi.org/10.1016/j.bbabio.2013.11.002
Dieteren CE, Willems PH, Vogel RO et al (2008) Subunits of mitochondrial complex I exist as part of matrix- and membrane-associated subcomplexes in living cells. J Biol Chem 283(50):34753–34761. https://doi.org/10.1074/jbc.M807323200
McKenzie M, Ryan MT (2010) Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life 62(7):497–502. https://doi.org/10.1002/iub.335
Guerrero-Castillo S, Baertling F, Kownatzki D et al (2017) The assembly pathway of mitochondrial respiratory chain complex I. Cell Metab 25(1):128–139. https://doi.org/10.1016/j.cmet.2016.09.002
Vogel RO, Janssen RJ, Ugalde C et al (2005) Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS J 272(20):5317–5326. https://doi.org/10.1111/j.1742-4658.2005.04928.x
Nouws J, Nijtmans L, Houten SM et al (2010) Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab 12(3):283–294. https://doi.org/10.1016/j.cmet.2010.08.002
Lazarou M, Thorburn DR, Ryan MT et al (2009) Assembly of mitochondrial complex I and defects in disease. Biochim Biophys Acta 1793(1):78–88. https://doi.org/10.1016/j.bbamcr.2008.04.015
Heide H, Bleier L, Steger M et al (2012) Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab 16(4):538–549. https://doi.org/10.1016/j.cmet.2012.08.009
Andrews B, Carroll J, Ding S et al (2013) Assembly factors for the membrane arm of human complex I. Proc Natl Acad Sci USA 110(47):18934–18939. https://doi.org/10.1073/pnas.1319247110
Guarani V, Paulo J, Zhai B et al (2014) TIMMDC1/C3orf1 functions as a membrane-embedded mitochondrial complex I assembly factor through association with the MCIA complex. Mol Cell Biol 34(5):847–861. https://doi.org/10.1128/mcb.01551-13
Hayashi T, Asano Y, Shintani Y et al (2015) Higd1a is a positive regulator of cytochrome c oxidase. Proc Natl Acad Sci USA 112(5):1553–1558. https://doi.org/10.1073/pnas.1419767112
Vidoni S, Harbour ME, Guerrero-Castillo S et al (2017) MR-1S interacts with PET100 and PET117 in module-based assembly of human cytochrome c oxidase. Cell Rep 18(7):1727–1738. https://doi.org/10.1016/j.celrep.2017.01.044
Vukotic M, Oeljeklaus S, Wiese S et al (2012) Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab 15(3):336–347. https://doi.org/10.1016/j.cmet.2012.01.016
Tausendschon M, Rehli M, Dehne N et al (1849) Genome-wide identification of hypoxia-inducible factor-1 and -2 binding sites in hypoxic human macrophages alternatively activated by IL-10. Biochim Biophys Acta 1:10–22. https://doi.org/10.1016/j.bbagrm.2014.10.006
Guo J, Chakraborty A, Liu P et al (2016) pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science 353(6302):929–932
Haack TB, Danhauser K, Haberberger B et al (2010) Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat Genet 42(12):1131–1134. https://doi.org/10.1038/ng.706
Dunning CJ, McKenzie M, Sugiana C et al (2007) Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J 26(13):3227–3237. https://doi.org/10.1038/sj.emboj.7601748
Nouws J, Nijtmans LG, Smeitink JA et al (2012) Assembly factors as a new class of disease genes for mitochondrial complex I deficiency. Cause, pathology and treatment options. Brain 135(Pt 1):12–22. https://doi.org/10.1093/brain/awr261
Taylor CT (2008) Mitochondria and cellular oxygen sensing in the HIF pathway. Biochem J 409(1):19–26. https://doi.org/10.1042/bj20071249
Vaupel P (2004) The role of hypoxia-induced factors in tumor progression. Oncologist 9(Suppl 5):10–17. https://doi.org/10.1634/theoncologist.9-90005-10
Fukuda R, Zhang H, Kim JW et al (2007) HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129(1):111–122. https://doi.org/10.1016/j.cell.2007.01.047
Chandel NS (2015) Evolution of mitochondria as signaling organelles. Cell Metab 22(2):204–206. https://doi.org/10.1016/j.cmet.2015.05.013
El Kasmi KC, Stenmark KR (2015) Contribution of metabolic reprogramming to macrophage plasticity and function. Semin Immunol 27(4):267–275. https://doi.org/10.1016/j.smim.2015.09.001
Tan Z, Xie N, Cui H et al (2015) Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J Immunol 194(12):6082–6089. https://doi.org/10.4049/jimmunol.1402469
Tannahill GM, Iraci N, Gaude E et al (2015) Metabolic reprogramming of mononuclear phagocytes in progressive multiple sclerosis. Front Immunol 6:106. https://doi.org/10.3389/fimmu.2015.00106
Biswas SK (2015) Metabolic reprogramming of immune cells in cancer progression. Immunity 43(3):435–449. https://doi.org/10.1016/j.immuni.2015.09.001
Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer metabolism. Cell Metab 23(1):27–47. https://doi.org/10.1016/j.cmet.2015.12.006
Jin Z, Wei W, Yang M et al (2014) Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab 20(3):483–498. https://doi.org/10.1016/j.cmet.2014.07.011
Wittig I, Braun HP, Schagger H (2006) Blue native PAGE. Nat Protoc 1(1):418–428. https://doi.org/10.1038/nprot.2006.62
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26(12):1367–1372. https://doi.org/10.1038/nbt.1511
Giese H, Ackermann J, Heide H et al (2015) NOVA. A software to analyze complexome profiling data. Bioinformatics 31(3):440–441. https://doi.org/10.1093/bioinformatics/btu623
Schwanhausser B, Busse D, Li N et al (2011) Global quantification of mammalian gene expression control. Nature 473(7347):337–342. https://doi.org/10.1038/nature10098
Wirth C, Brandt U, Hunte C et al (2016) Structure and function of mitochondrial complex I. Biochim Biophys Acta. https://doi.org/10.1016/j.bbabio.2016.02.013
Moreno-Lastres D, Fontanesi F, Garcia-Consuegra I et al (2012) Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab 15(3):324–335. https://doi.org/10.1016/j.cmet.2012.01.015
Acknowledgements
We thank Tanja Keppler for excellent technical assistance, Jana Meisterknecht for competent support with BNE and 2D-BNE/SDS gels, and Ilka Siebels for technical assistance in respiration measurements.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft [SFB 815, project Z1 (I.W.) and project A8 (B.B.)].
Author information
Authors and Affiliations
Contributions
DF performed and planed the experiments and wrote the paper. IW performed complexome profiling and participated on paper writing. SD performed respiratory measurements. TS gave his expertise concerning β-TrCP experiments ND discussed data and literature. BB performed data interpretation, wrote the paper, and designed the project.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fuhrmann, D.C., Wittig, I., Dröse, S. et al. Degradation of the mitochondrial complex I assembly factor TMEM126B under chronic hypoxia. Cell. Mol. Life Sci. 75, 3051–3067 (2018). https://doi.org/10.1007/s00018-018-2779-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2779-y