Abstract
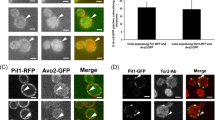
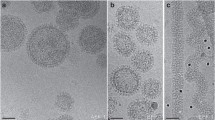
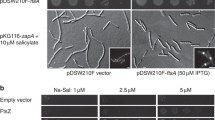
Dni1 and Dni2 facilitate cell fusion during mating. Here, we show that these proteins are interdependent for their localization in a plasma membrane subdomain, which we have termed the mating fusion domain. Dni1 compartmentation in the domain is required for cell fusion. The contribution of actin, sterol-dependent membrane organization, and Dni2 to this compartmentation was analysed, and the results showed that Dni2 plays the most relevant role in the process. In turn, the Dni2 exit from the endoplasmic reticulum depends on Dni1. These proteins share the presence of a cysteine motif in their first extracellular loop related to the claudin GLWxxC(8–10 aa)C signature motif. Structure–function analyses show that mutating each Dni1 conserved cysteine has mild effects, and that only simultaneous elimination of several cysteines leads to a mating defect. On the contrary, eliminating each single cysteine and the C-terminal tail in Dni2 abrogates Dni1 compartmentation and cell fusion. Sequence alignments show that claudin trans-membrane helixes bear small-XXX-small motifs at conserved positions. The fourth Dni2 trans-membrane helix tends to form homo-oligomers in Escherichia plasma membrane, and two concatenated small-XXX-small motifs are required for efficient oligomerization and for Dni2 export from the yeast endoplasmic reticulum. Together, our results strongly suggest that Dni2 is an ancient claudin that blocks Dni1 diffusion from the intercellular region where two plasma membranes are in close proximity, and that this function is required for Dni1 to facilitate cell fusion.







Similar content being viewed by others
Abbreviations
- CAT:
-
Chloramphenicol acetyl transferase
- ECL:
-
Extra-cellular loop
- ER:
-
Endoplasmic reticulum
- GFP:
-
Green fluorescent protein
- MFD:
-
Membrane fusion domain
- PDZ:
-
Post-synaptic density protein PSD95, Drosophila disc large tumour suppressor Dlg1, and zonula occludens-1 zo-1 proteins
- TJ:
-
Tight junction
- TMD:
-
Trans-membrane Domain
- WT:
-
Wild-type
References
Chen EH, Grote E, Mohler W, Vignery A (2007) Cell-cell fusion. FEBS Lett 581(11):2181–2193
Aguilar PS, Baylies MK, Fleissner A, Helming L, Inoue N, Podbilewicz B, Wang H, Wong M (2013) Genetic basis of cell-cell fusion mechanisms. Trends Genet 29(7):427–437
Ydenberg CA, Rose MD (2008) Yeast mating: a model system for studying cell and nuclear fusion. Methods Mol Biol (Clifton, NJ) 475:3–20
Merlini L, Dudin O, Martin SG (2013) Mate and fuse: how yeast cells do it. Open Biol 3(3):130008
Martin SG (2016) Role and organization of the actin cytoskeleton during cell-cell fusion. Semin Cell Dev Biol 60:121–126
White JM, Rose MD (2001) Yeast mating: getting close to membrane merger. Curr Biol 11(1):R16–R20
Heiman MG, Walter P (2000) Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol 151:719–730
Fleissner A, Diamond S, Glass NL (2009) The Saccharomyces cerevisiae PRM1 homolog in Neurospora crassa is involved in vegetative and sexual cell fusion events but also has postfertilization functions. Genetics 181(2):497–510
Curto MA, Sharifmoghadam MR, Calpena E, De Leon N, Hoya M, Doncel C, Leatherwood J, Valdivieso MH (2014) Membrane organization and cell fusion during mating in fission yeast requires multipass membrane protein Prm1. Genetics 196(4):1059–1076
Clemente-Ramos JA, Martin-Garcia R, Sharifmoghadam MR, Konomi M, Osumi M, Valdivieso MH (2009) The tetraspan protein Dni1p is required for correct membrane organization and cell wall remodelling during mating in Schizosaccharomyces pombe. Mol Microbiol 73(4):695–709
Muller EM, Mackin NA, Erdman SE, Cunningham KW (2003) Fig 1p facilitates Ca2+ influx and cell fusion during mating of Saccharomyces cerevisiae. J Biol Chem 278:38461–38469
Proszynski TJ, Klemm R, Bagnat M, Gaus K, Simons K (2006) Plasma membrane polarization during mating in yeast cells. J Cell Biol 173(6):861–866
Bagnat M, Simons K (2002) Cell surface polarization during yeast mating. Proc Natl Acad Sci USA 99(22):14183–14188
Olmo VN, Grote E (2010) Prm1 targeting to contact sites enhances fusion during mating in Saccharomyces cerevisiae. Eukaryot Cell 9(10):1538–1548
Anderson RG, Jacobson K (2002) A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296(5574):1821–1825
Laude AJ, Prior IA (2004) Plasma membrane microdomains: organization, function and trafficking. Mol Membr Biol 21(3):193–205
Garcia-Parajo MF, Cambi A, Torreno-Pina JA, Thompson N, Jacobson K (2014) Nanoclustering as a dominant feature of plasma membrane organization. J Cell Sci 127(Pt 23):4995–5005
Corcoran JA, Salsman J, de Antueno R, Touhami A, Jericho MH, Clancy EK, Duncan R (2006) The p14 fusion-associated small transmembrane (FAST) protein effects membrane fusion from a subset of membrane microdomains. J Biol Chem 281(42):31778–31789
Olivera-Couto A, Aguilar PS (2012) Eisosomes and plasma membrane organization. Mol Genet Genom 287(8):607–620
Roth MG (2004) Phosphoinositides in constitutive membrane traffic. Physiol Rev 84(3):699–730
Rubinstein E (2011) The complexity of tetraspanins. Biochem Soc Trans 39(2):501–505
Trimble WS, Grinstein S (2015) Barriers to the free diffusion of proteins and lipids in the plasma membrane. J Cell Biol 208(3):259–271
Valdez-Taubas J, Pelham HR (2003) Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr Biol 13(18):1636–1640
Balda MS, Matter K (2016) Tight junctions as regulators of tissue remodelling. Curr Opin Cell Biol 42:94–101
Lahiri S, Toulmay A, Prinz WA (2015) Membrane contact sites, gateways for lipid homeostasis. Curr Opin Cell Biol 33:82–87
Simons K, Fuller SD (1985) Cell surface polarity in epithelia. Annu Rev Cell Biol 1:243–288
Gunzel D, Yu AS (2013) Claudins and the modulation of tight junction permeability. Physiol Rev 93(2):525–569
Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM (2007) Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446(7137):801–805
Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S (1998) Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol 141(7):1539–1550
Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE (2008) Structure and function of claudins. Biochem Biophys Acta 1778(3):631–645
Krause G, Protze J, Piontek J (2015) Assembly and function of claudins: structure-function relationships based on homology models and crystal structures. Semin Cell Dev Biol 42:3–12
Lingaraju A, Long TM, Wang Y, Austin JR 2nd, Turner JR (2015) Conceptual barriers to understanding physical barriers. Semin Cell Dev Biol 42:13–21
Aguilar PS, Engel A, Walter P (2007) The plasma membrane proteins Prm1 and Fig 1 ascertain fidelity of membrane fusion during yeast mating. Mol Biol Cell 18(2):547–556
Alvarez FJ, Douglas LM, Rosebrock A, Konopka JB (2008) The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol Biol Cell 19(12):5214–5225
Forsburg SL, Rhind N (2006) Basic methods for fission yeast. Yeast 23(3):173–183
Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194:795–823
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. CSHL Press, New York
Kunkel TA, Roberts JD, Zakour RA (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol 154:367–382
Maundrell K (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127–130
Stern B, Nurse P (1997) Fission yeast pheromone blocks S-phase by inhibiting the G1 cyclinB-p34cdc2 kinase. EMBO J 16:534
Sharifmoghadam MR, Valdivieso MH (2009) The fission yeast SEL1 domain protein Cfh3p: a novel regulator of the glucan synthase Bgs1p whose function is more relevant under stress conditions. J Biol Chem 284(17):11070–11079
de Leon N, Hoya M, Curto MA, Moro S, Yanguas F, Doncel C, Valdivieso MH (2016) The AP-2 complex is required for proper temporal and spatial dynamics of endocytic patches in fission yeast. Mol Microbiol 100(3):409–424
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Russ WP, Engelman DM (1999) TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci USA 96(3):863–868
Armstrong CR (1858) Senes A (2016) Screening for transmembrane association in divisome proteins using TOXGREEN, a high-throughput variant of the TOXCAT assay. Biochem Biophys Acta 11:2573–2583
Dudin O, Bendezu FO, Groux R, Laroche T, Seitz A, Martin SG (2015) A formin-nucleated actin aster concentrates cell wall hydrolases for cell fusion in fission yeast. J Cell Biol 208(7):897–911
Lock A, Forfar R, Weston C, Bowsher L, Upton GJ, Reynolds CA, Ladds G, Dixon AM (2014) One motif to bind them: a small-XXX-small motif affects transmembrane domain 1 oligomerization, function, localization, and cross-talk between two yeast GPCRs. Biochem Biophys Acta 1838(12):3036–3051
Hsin J, LaPointe LM, Kazy A, Chipot C, Senes A, Schulten K (2011) Oligomerization state of photosynthetic core complexes is correlated with the dimerization affinity of a transmembrane helix. J Am Chem Soc 133(35):14071–14081
Senes A, Gerstein M, Engelman DM (2000) Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol 296(3):921–936
Gong Y, Renigunta V, Zhou Y, Sunq A, Wang J, Yang J, Renigunta A, Baker LA, Hou J (2015) Biochemical and biophysical analyses of tight junction permeability made of claudin-16 and claudin-19 dimerization. Mol Biol Cell 26(24):4333–4346
Rossa J, Ploeger C, Vorreiter F, Saleh T, Protze J, Gunzel D, Wolburg H, Krause G, Piontek J (2014) Claudin-3 and claudin-5 protein folding and assembly into the tight junction are controlled by non-conserved residues in the transmembrane 3 (TM3) and extracellular loop 2 (ECL2) segments. J Biol Chem 289(11):7641–7653
Teese MG, Langosch D (2015) Role of GxxxG motifs in transmembrane domain interactions. Biochemistry 54(33):5125–5135
Koval M (2013) Differential pathways of claudin oligomerization and integration into tight junctions. Tissue Barriers 1(3):e24518
Iwaki T, Tanaka N, Takagi H, Giga-Hama Y, Takegawa K (2004) Characterization of end4+, a gene required for endocytosis in Schizosaccharomyces pombe. Yeast 21(10):867–881
Wachtler V, Rajagopalan S, Balasubramanian MK (2003) Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J Cell Sci 116(Pt 5):867–874
Krug SM, Schulzke JD, Fromm M (2014) Tight junction, selective permeability, and related diseases. Semin Cell Dev Biol 36:166–176
Simske JS (2013) Claudins reign: the claudin/EMP/PMP22/gamma channel protein family in C. elegans. Tissue Barriers 1(3):25502
Hou J, Renigunta A, Gomes AS, Hou M, Paul DL, Waldegger S, Goodenough DA (2009) Claudin-16 and claudin-19 interaction is required for their assembly into tight junctions and for renal reabsorption of magnesium. Proc Natl Acad Sci USA 106(36):15350–15355
Zihni C, Mills C, Matter K, Balda MS (2016) Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev 17(9):564–580
Lambert D, O’Neill CA, Padfield PJ (2007) Methyl-beta-cyclodextrin increases permeability of Caco-2 cell monolayers by displacing specific claudins from cholesterol rich domains associated with tight junctions. Cell Physiol Biochem 20(5):495–506
Krause SA, Xu H, Gray JV (2008) The synthetic genetic network around PKC1 identifies novel modulators and components of protein kinase C signaling in Saccharomyces cerevisiae. Eukaryot Cell 7(11):1880–1887
Markvoort AJ, Marrink SJ (2011) Lipid acrobatics in the membrane fusion arena. Curr Top Membr 68:259–294
Lee DB, Jamgotchian N, Allen SG, Abeles MB, Ward HJ (2008) A lipid-protein hybrid model for tight junction. Am J Physiol Renal Physiol 295(6):F1601–F1612
Nguyen VS, Jobichen C, Tan KW, Tan YW, Chan SL, Ramesh K, Yuan Y, Hong Y, Seetharaman J, Leung KY, Sivaraman J, Mok YK (2015) Structure of AcrH-AopB chaperone-translocator complex reveals a role for membrane hairpins in type III secretion system translocon assembly. Structure 23(11):2022–2031
Hemler ME (2008) Targeting of tetraspanin proteins–potential benefits and strategies. Nat Rev Drug Discov 7(9):747–758
Sourisseau M, Michta ML, Zony C, Israelow B, Hopcraft SE, Narbus CM, Parra Martin A, Evans MJ (2013) Temporal analysis of hepatitis C virus cell entry with occludin directed blocking antibodies. PLoS Pathog 9(3):e1003244
Acknowledgements
We thank E. Keck for the English revision. We are indebted to S. Moreno, D. Mulvihill, O. Nielsen, P. Nurse, P. Pérez, J.C. Ribas, Y. Sanchez, C. Shimoda, and K. Takegawa for strains and plasmids. A. Senes and C. Armstrong are acknowledged for providing plasmids and the guidance to perform the TOXCAT analyses. Financial support from the Ministerio de Economía y Competitividad (MINECO, Spain)/European Union FEDER program (BFU2013-48582-C2-2-P) and Junta de Castilla y León (Grant SA073U14) made this work possible. MAC, SM, and FY were supported by FPU fellowships from the Ministerio de Educación. CGG was supported by the Sistema de Garantía Juvenil program from the Ministerio de Empleo y Seguridad Social, Spain.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Curto, MÁ., Moro, S., Yanguas, F. et al. The ancient claudin Dni2 facilitates yeast cell fusion by compartmentalizing Dni1 into a membrane subdomain. Cell. Mol. Life Sci. 75, 1687–1706 (2018). https://doi.org/10.1007/s00018-017-2709-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-017-2709-4