Abstract
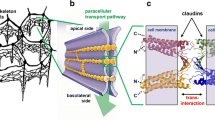
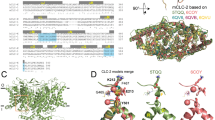
Claudin-17 is a paracellular channel-forming tight junction protein. Unlike the cation channels claudin-2 and -15, claudin-17 forms a distinct anion-selective channel. Aim of this study was to determine the molecular basis of channel formation and charge selectivity of this protein. To achieve this, residues located in the extracellular loops (ECL) 1 and 2 of claudin-17 were substituted, preferably those whose charges differed in claudin-17 and in claudin-2 or -15. The respective mutants were stably expressed in MDCK C7 cells and their ability to form charge-selective channels was analyzed by measuring ion permeabilities and transepithelial electrical resistance. The functional data were combined with homology modeling of the claudin-17 protomer using the structure of claudin-15 as template. In ECL1, K65, R31, E48, and E44 were found to be stronger involved in Cldn17 channel function than the clustered R45, R56, R59, and R61. For K65, not only charge but also stereochemical properties were crucial for formation of the anion-selective channel. In ECL2, both Y149 and H154 were found to contribute to constitution of the anion channel in a distinct manner. In conclusion, we provide insight into the molecular mechanism of the formation of charge- and size-selective paracellular ion channels. In detail, we propose a hydrophilic furrow in the claudin-17 protomer spanning from a gap between the ends of TM2 and TM3 along R31, E48, and Y67 to a gap between K65 and S68 lining the anion channel.







Similar content being viewed by others
Abbreviations
- TJ:
-
Tight junction
- Cldn:
-
Claudin
- Occl:
-
Occludin
- SEM:
-
Standard error of the mean
- MDCK:
-
Madin-Darby Canine Kidney
- TER:
-
Transepithelial resistance
- wt:
-
Wildtype
- vec:
-
Vector control
- TM:
-
Transmembrane segment
- ECL1 and -2:
-
Extracellular loop 1 and 2, equivalent to extracellular segment 1 and 2
References
Krause G et al (2008) Structure and function of claudins. Biochim Biophys Acta 1778(3):631–645
Colegio OR et al (2003) Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am J Physiol Cell Physiol 284(6):C1346–C1354
Li J et al (2013) Conserved aromatic residue confers cation selectivity in claudin-2 and claudin-10b. J Biol Chem 288(31):22790–22797
Günzel D, Fromm M (2012) Claudins and other tight junction proteins. Compr Physiol 2(3):1819–1852
Mineta K et al (2011) Predicted expansion of the claudin multigene family. FEBS Lett 585(4):606–612
Krug SM, Schulzke JD, Fromm M (2014) Tight junction, selective permeability, and related diseases. Semin Cell Dev Biol 36:166–176
Amasheh S et al (2002) Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J Cell Sci 115(Pt 24):4969–4976
Van Itallie CM et al (2006) Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am J Physiol Renal Physiol 291(6):F1288–F1299
Günzel D et al (2009) Claudin-10 exists in six alternatively spliced isoforms that exhibit distinct localization and function. J Cell Sci 122(Pt 10):1507–1517
Van Itallie CM, Fanning AS, Anderson JM (2003) Reversal of charge selectivity in cation or anion-selective epithelial lines by expression of different claudins. Am J Physiol Renal Physiol 285(6):F1078–F1084
Krug SM et al (2012) Claudin-17 forms tight junction channels with distinct anion selectivity. Cell Mol Life Sci 69(16):2765–2778
Rosenthal R et al (2010) Claudin-2, a component of the tight junction, forms a paracellular water channel. J Cell Sci 123(Pt 11):1913–1921
Angelow S, Yu AS (2009) Structure-function studies of claudin extracellular domains by cysteine-scanning mutagenesis. J Biol Chem 284(42):29205–29217
Yu AS et al (2009) Molecular basis for cation selectivity in claudin-2-based paracellular pores: identification of an electrostatic interaction site. J Gen Physiol 133(1):111–127
Wen H et al (2004) Selective decrease in paracellular conductance of tight junctions: role of the first extracellular domain of claudin-5. Mol Cell Biol 24(19):8408–8417
Piehl C et al (2010) Participation of the second extracellular loop of claudin-5 in paracellular tightening against ions, small and large molecules. Cell Mol Life Sci 67(12):2131–2140
Piontek J et al (2008) Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J 22(1):146–158
Veshnyakova A et al (2012) Determinants contributing to claudin ion channel formation. Ann N Y Acad Sci 1257:45–53
Suzuki H et al (2014) Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 344(6181):304–307
Saitoh Y et al (2015) Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science 347(6223):775–778
Biasini M et al (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42((Web server issue)):W252–W258
Bordoli L et al (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4(1):1–13
Arnold K et al (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22(2):195–201
Benkert P, Kunzli M, Schwede T (2009) QMEAN server for protein model quality estimation. Nucleic Acids Res 37((Web Server issue)):W510–W514
Suzuki et al (2015) Model for the architecture of claudin-based paracellular ion channels through tight junctions. J Mol Biol 427(2):291–297
Hou J et al (2010) Claudin-4 forms paracellular chloride channel in the kidney and requires claudin-8 for tight junction localization. Proc Natl Acad Sci USA 107(42):18010–18015
Colegio OR et al (2002) Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol 283(1):C142–C147
Konrad M et al (2006) Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet 79(5):949–957
Krause G, Protze J, Piontek J (2015) Assembly and function of claudins: Structure-function relationships based on homology models and crystal structures. Semin Cell Dev Biol. doi:10.1016/j.semcdb.2015.04.010 [Epub ahead of print]
Alexandre MD et al (2007) The first extracellular domain of claudin-7 affects paracellular Cl- permeability. Biochem Biophys Res Commun 357(1):87–91
Hou J, Paul DL, Goodenough DA (2005) Paracellin-1 and the modulation of ion selectivity of tight junctions. J Cell Sci 118(Pt 21):5109–5118
Kausalya PJ et al (2006) Disease-associated mutations affect intracellular traffic and paracellular Mg2+ transport function of Claudin-16. J Clin Invest 116(4):878–891
Rossa J et al (2014) Molecular and structural transmembrane determinants critical for embedding claudin-5 into tight junctions reveal a distinct four-helix bundle arrangement. Biochem J 464(1):49–60
Protze J et al (2015) Directed structural modification of Clostridium perfringens enterotoxin to enhance binding to claudin-5. Cell Mol Life Sci 72(7):1417–1432
Winkler L et al (2009) Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J Biol Chem 284(28):18863–18872
Rossa J et al (2014) Claudin-3 and claudin-5 protein folding and assembly into the tight junction are controlled by non-conserved residues in the transmembrane 3 (TM3) and extracellular loop 2 (ECL2) segments. J Biol Chem 289(11):7641–7653
Piontek J et al (2011) Elucidating the principles of the molecular organization of heteropolymeric tight junction strands. Cell Mol Life Sci 68(23):3903–3918
Van Itallie CM, Mitic LL, Anderson JM (2011) Claudin-2 forms homodimers and is a component of a high molecular weight protein complex. J Biol Chem 286(5):3442–3450
Acknowledgments
We appreciate the excellent technical assistance of In-Fah M. Lee and Detlef Sorgenfrei. This study was supported by grants of the Deutsche Forschungsgemeinschaft (DFG FOR 721).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
M. P. Conrad and J. Piontek contributed equally to the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2015_1987_MOESM1_ESM.tiff
Figure S1. Multiple sequence alignment of ECL1 (left) and ECL2 (right) for Cldn17. 28 orthologous sequences available in UNIPROTKB were aligned using CLUSTAL O (McWilliam et al., Nucleic Acids Res. 2013 Jul; 41(Web Server issue: W597-600.)). Accession number and species is given on the left; residue number is given on top; green, positively charged; red negatively charged; cyan Ser/Thr; *, identical; :, strongly similar properties;., weakly similar properties. The sequences are shown with positions 29 - 81 and 146-164 (ECL1 and ECL2 as defined by uniprot.org). Boxes mark the residues analyzed in this study. Blue boxes mark positions with identical or highly similar residues whereas red boxes mark positions with residues containing different properties (TIFF 2337 kb)
18_2015_1987_MOESM2_ESM.tif
Figure S2. Exemplary western blots for other TJ proteins. The blots show exemplary clones of each mutation in comparison to wt Cldn17 and vector control (vec). Detected were Cldn1, Cldn4, occludin, the FLAG-tagged Cldn17 and β-actin as loading control. Expression changes occurred in some, but not all, clones of one mutant. However, these changes had no impact on functional results, as these were similar in all clones of one mutant (TIFF 221 kb)
18_2015_1987_MOESM3_ESM.tiff
Figure S3. Exemplary comparison of different clones (E44A). a The blots show exemplary clones of mutation E44A in comparison to wt Cldn17 and a vector control (vec). Detected were Cldn1, Cldn4, occludin, the FLAG-tagged Cldn17 and β-actin as loading control. b The densitometric analysis revealed expression differences which may influence the results of further experiments. However, only a very low mutant expression of 14 % (clone #1) compared to the wt, resulted in PCl/PNa as observed for the vec, while ranges of expression above 43 % to 86 % were comparable to each other c. Also permeabilities for other ions d were only different from the other clones at very low expression levels. Additional expression variations in endogenous TJ protein levels had no effect in the ranges observed. However, to keep clonal variation as low as possible, several clones were analyzed for each mutant and outliers with extreme differences in expressions were excluded (TIFF 3315 kb)
18_2015_1987_MOESM4_ESM.tiff
Figure S4. Immunofluorescent staining of exemplary clones. Localization of the 3 × FLAG-tagged mutants of Cldn17 (red) was analyzed using occludin or ZO-1 (green) as TJ marker. All mutant Cldn17 constructs were colocalized (yellow) with the TJ marker, indicating correct insertion into the TJ. Bar = 10 µm (TIFF 16234 kb)
18_2015_1987_MOESM5_ESM.tiff
Figure S5. Exemplary images of freeze-fracture electron microscopic samples. Mutation from anion- to cation-selective Cldn17 (mutant K65E) as well as the other mutations of K65 that led to loss of charge selectivity had no effect on TJ ultrastructure. a Vector-transfected control. b wt Cldn17. c Cldn17 K65E. d Cldn17 K65A. e Cldn17 K65R. Bar = 200 nm (TIFF 4714 kb)
18_2015_1987_MOESM6_ESM.tiff
Figure S6. Live cell imaging of HEK293 cells transfected with three CFP-tagged constructs. Subcellular localization of a wtCldn17, b Cldn17 Y149A and c Cldn17 H154A (green). Trypan blue (red) marks the cell membrane. All three Cldn17 constructs were expressed and localized within the cell membrane. Enrichment in cell membranes of neighboring cells indicates the ability to trans-interact. Bar 5 µm (TIFF 20632 kb)
18_2015_1987_MOESM7_ESM.tiff
Figure S7. Overlay of Cldn17 and Cldn19 homology models. Comparison of Cldn17 homology models based on templates for Cldn15 (PDB: 4P79, backbone as cartoon in green, residues as sticks in magenta) and Cldn19 (PDB: 3X29, backbone as cartoon in cyan, residues as sticks in violet). Both models indicate a similar fold of Cldn17 and similar positions of the residues (sticks) mutated in this study. However, the models differ in the C-terminal half of ECL2, the loop between β1- and β2-strand and the region between β4-strand and TM2. The latter region was not resolved in the Cldn19 crystal, in which Cldn19 was in complex with the C-terminal domain of the Clostridium perfringens enterotoxin [20]. Disulfide bridges in ECL1 are shown as yellow sticks (TIFF 2927 kb)
Rights and permissions
About this article
Cite this article
Conrad, M.P., Piontek, J., Günzel, D. et al. Molecular basis of claudin-17 anion selectivity. Cell. Mol. Life Sci. 73, 185–200 (2016). https://doi.org/10.1007/s00018-015-1987-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-015-1987-y